25.3 DNA Recombination
The rearrangement of genetic information within and among DNA molecules encompasses a variety of processes, collectively placed under the heading of genetic recombination. The practical applications of DNA rearrangements in altering the genomes of increasing numbers of organisms are now being explored (Chapter 9).

Barbara McClintock, 1902–1992
Genetic recombination events fall into at least three general classes. Homologous genetic recombination (also called general recombination) involves genetic exchanges between any two DNA molecules (or segments of the same molecule) that share an extended region of nearly identical sequence. The actual sequence of bases is irrelevant, as long as it is similar in the two DNAs. In site-specific recombination, the exchanges occur only at a particular DNA sequence. DNA transposition is distinct from both other classes in that it usually involves a short segment of DNA with the remarkable capacity to move from one location in a chromosome to another. These “jumping genes” were first observed in maize in the 1940s by Barbara McClintock. There are also unusual genetic rearrangements for which no mechanism or purpose has yet been proposed. Here we focus on the three general classes.
Homologous genetic recombination is largely a pathway to repair double-strand breaks in DNA. An alternative process for double-strand break repair that does not entail recombination, called nonhomologous end joining (NHEJ), is also described here. Genetic recombination systems have functions as varied as their mechanisms. They include roles in specialized DNA repair systems, specialized activities in DNA replication, regulation of expression of certain genes, facilitation of proper chromosome segregation during eukaryotic cell division, maintenance of genetic diversity, and implementation of programmed genetic rearrangements during embryonic development. In most cases, genetic recombination is closely integrated with other processes in DNA metabolism, and this becomes a theme of our discussion.
Bacterial Homologous Recombination Is a DNA Repair Function
In bacteria, homologous genetic recombination is primarily a DNA repair process, and in this context (as noted in Section 25.2) it is referred to as recombinational DNA repair. It is usually directed at the reconstruction of replication forks that have stalled or collapsed at the site of DNA damage. Homologous genetic recombination can also occur during conjugation (mating), when chromosomal DNA is transferred from one bacterial cell (donor) to another (recipient). Recombination during conjugation, although rare in wild bacterial populations, contributes to genetic diversity.
When a replication fork encounters DNA damage, many pathways may resolve the conflict. A common feature of the DNA repair pathways illustrated in Figures 25-21 to 25-24 is that they introduce a transient break into one of the DNA strands. If a replication fork encounters a damaged site under repair near a break in one of the template strands, one arm of the replication fork becomes disconnected by a double-strand break and the fork collapses (Fig. 25-29). The end of that break is processed by degrading the -ending strand. The resulting single-stranded extension is bound by a recombinase that uses it to promote strand invasion: the end invades the intact duplex DNA connected to the other arm of the fork and pairs with its complementary sequence. This creates a branched DNA structure (a point where three DNA segments come together). The DNA branch can be moved in a process called branch migration to create an X-like crossover structure known as a Holliday intermediate, named after researcher Robin Holliday, who first postulated its existence. The Holliday intermediate is cleaved, or “resolved,” by a special class of nucleases. The overall process reconstructs the replication fork.
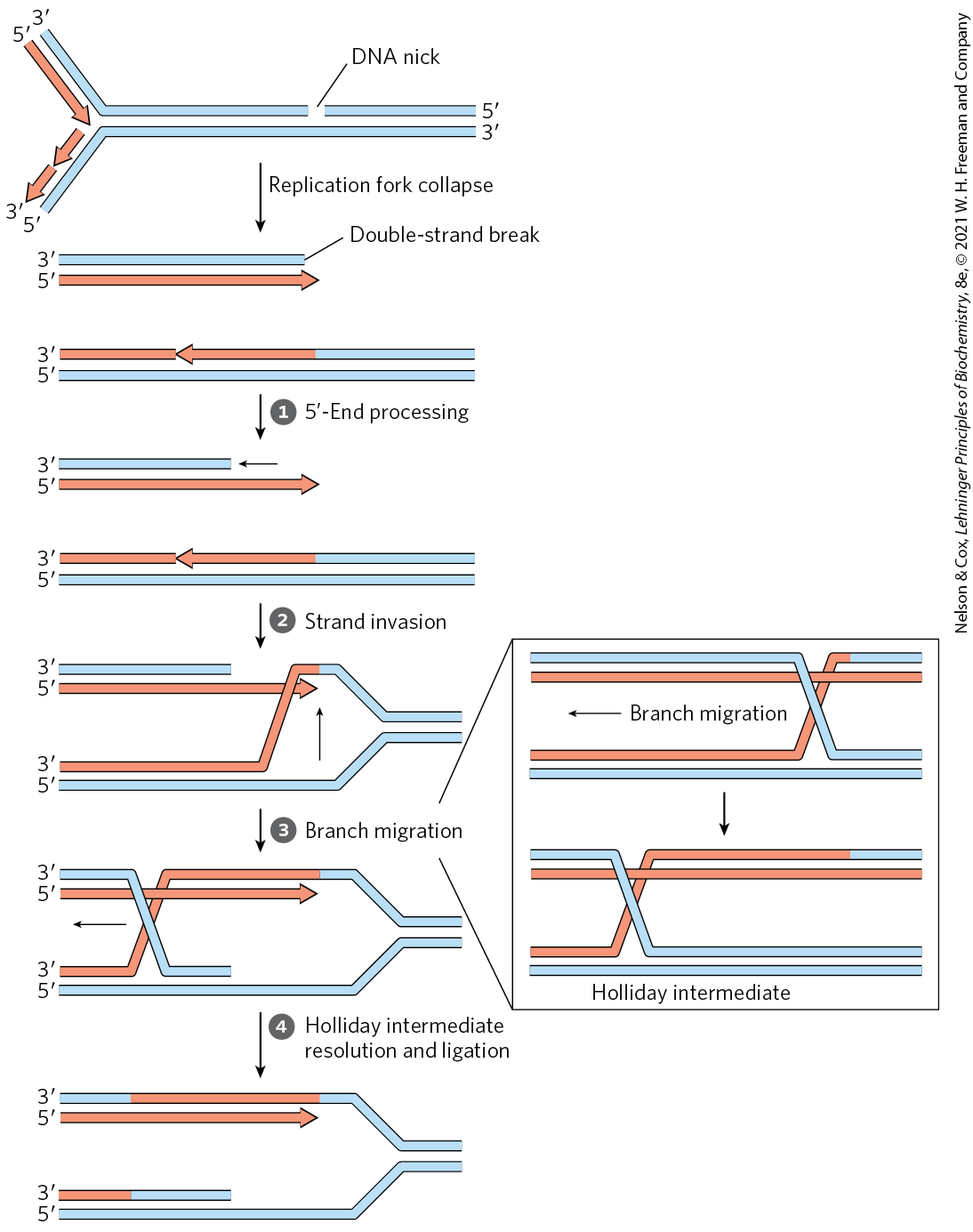
FIGURE 25-29 Recombinational DNA repair at a collapsed replication fork. When a replication fork encounters a break in one of the template strands, one arm of the fork is lost and the replication fork collapses. The -ending strand at the break is degraded to create a single-stranded extension, which is then used in a strand invasion process, pairing the invading single strand with its complementary strand within the adjacent duplex. Migration of the branch (shown in the box) can create a Holliday intermediate. Cleavage of the Holliday intermediate by specialized nucleases, followed by ligation, restores a viable replication fork. The replisome is reloaded onto this structure (not shown), and replication continues. Arrowheads represent ends.
In E. coli, the DNA end-processing is promoted by the RecBCD nuclease/helicase. The RecBCD enzyme binds to linear DNA at a free (broken) end and moves inward along the double helix, unwinding and degrading the DNA in a reaction coupled to ATP hydrolysis (Fig. 25-30). The RecB and RecD subunits are helicase motors, with RecB moving along one strand, and RecD moving along the other strand. The activity of the enzyme is altered when it interacts with a sequence referred to as chi, , which binds tightly to a site on the RecC subunit. From that point, degradation of the strand with a terminus is greatly reduced, but degradation of the -terminal strand is increased. This process creates a single-stranded DNA with a end, which is used during subsequent steps in recombination. The 1,009 chi sequences scattered throughout the E. coli genome enhance the frequency of recombination about 5- to 10-fold within 1,000 bp of each chi site. The enhancement declines as the distance from chi increases. Sequences that enhance recombination frequency have also been identified in several other organisms.
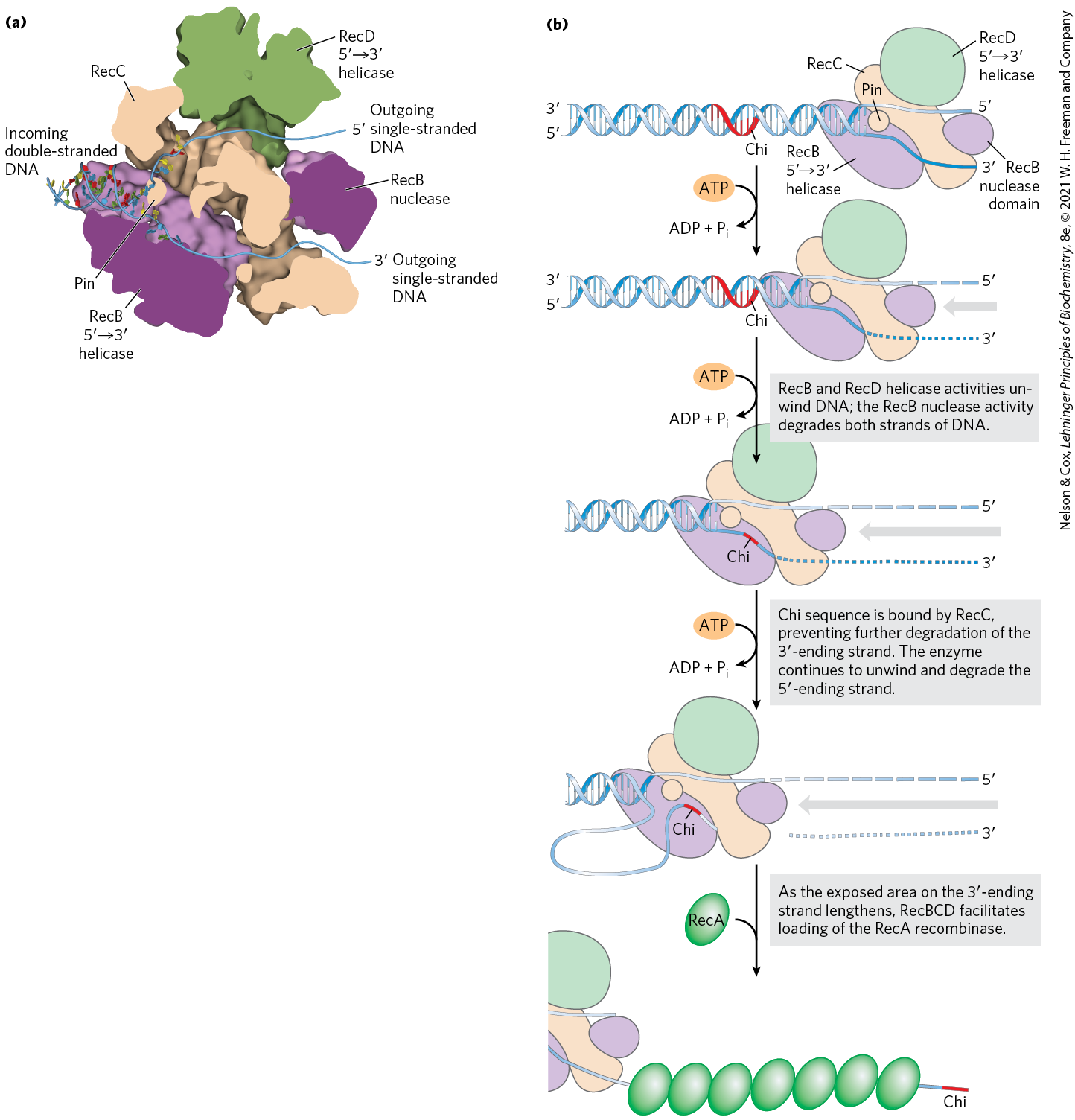
FIGURE 25-30 The RecBCD helicase/nuclease. (a) A cutaway view of the RecBCD enzyme structure as it is bound to DNA. The subunits are shown in different colors; the DNA is entering from the left, and the unwound DNA strands (not part of the solved structure) are shown exiting to the right. A bulbous protein structure called a pin, part of the RecC subunit, facilitates the separation of strands. (b) Activities of the RecBCD enzyme at a DNA end. [(a) Data from PDB ID 1W36, M. R. Singleton et al., Nature 432:187, 2004.]
The bacterial recombinase is the RecA protein. RecA is unusual among the proteins of DNA metabolism in that its active form is an ordered, helical filament of up to several thousand subunits that assemble cooperatively on DNA (Fig. 25-31). This filament usually forms on single-stranded DNA, such as that produced by the RecBCD enzyme. Its formation is not as straightforward as shown in Figure 25-31, because the single-stranded DNA–binding protein (SSB) is normally present and specifically impedes the binding of the first few subunits to DNA (filament nucleation). The RecBCD enzyme acts directly as a RecA loader, facilitating the nucleation of a RecA filament on single-stranded DNA that is coated with SSB. The filaments assemble and disassemble predominantly in a direction. Many other bacterial proteins regulate the formation and disassembly of RecA filaments, including an alternative set of RecA loading proteins called RecF, RecO, and RecR. RecA protein promotes the central steps of homologous recombination, including the DNA strand invasion step of Figure 25-29, as well as other strand exchange reactions occurring in vitro. Once a Holliday intermediate has been created via branch migration, it can be cleaved by specialized nucleases such as the bacterial RuvC protein (Fig. 25-32), and nicks are sealed by DNA ligase. A viable replication fork structure is thus reconstructed, as outlined in Figure 25-29.
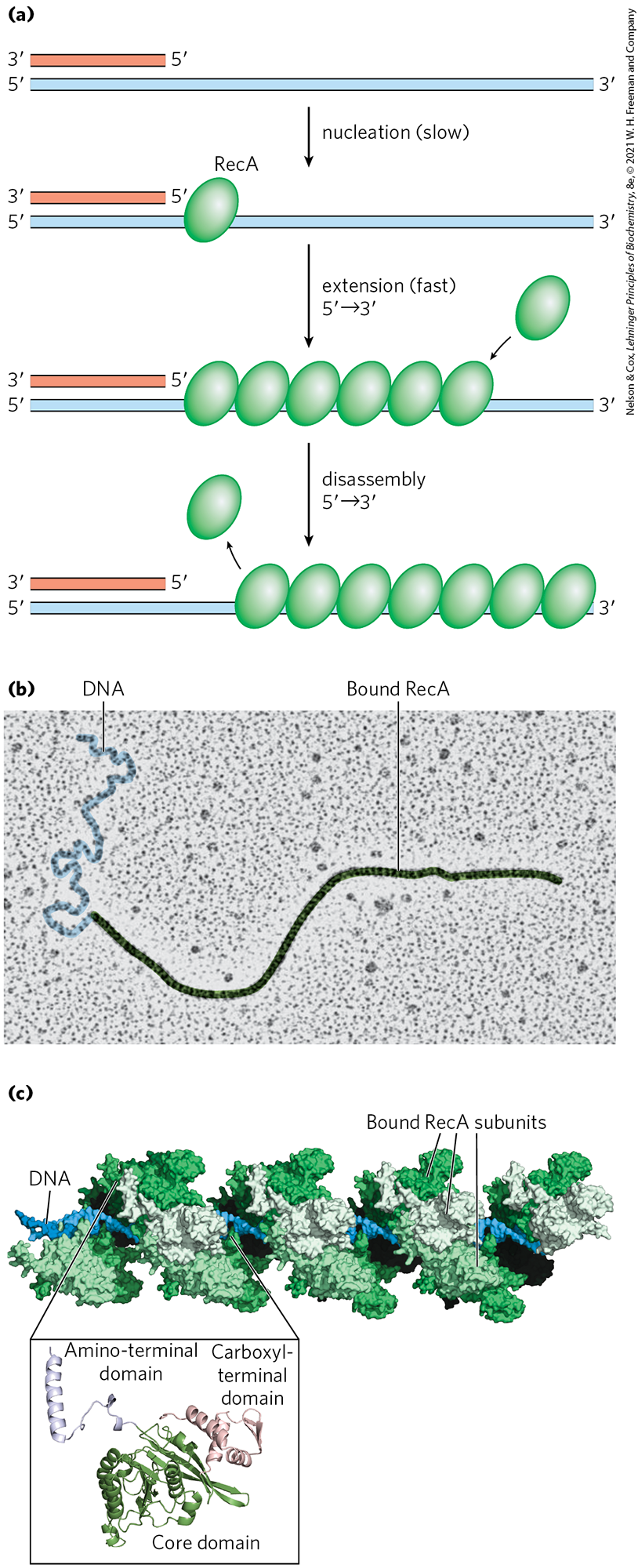
FIGURE 25-31 RecA protein filaments. RecA and other recombinases in this class function as filaments of nucleoprotein. (a) Filament formation proceeds in discrete nucleation and extension steps. Nucleation is the addition of the first few RecA subunits. Extension occurs by adding RecA subunits so that the filament grows in the direction. When disassembly occurs, subunits are subtracted from the trailing end. (b) Colorized electron micrograph of a RecA filament bound to DNA. (c) Segment of a RecA filament with four helical turns (24 RecA subunits). Notice the bound double-stranded DNA in the center. The core domain of RecA is structurally related to the motor domains of helicases. [(b) By permission of the Estate of Ross Inman. Special thanks to Kim Voss. (c) Data from PDB ID 3CMX, Z. Chen et al., Nature 453:489, 2008.]
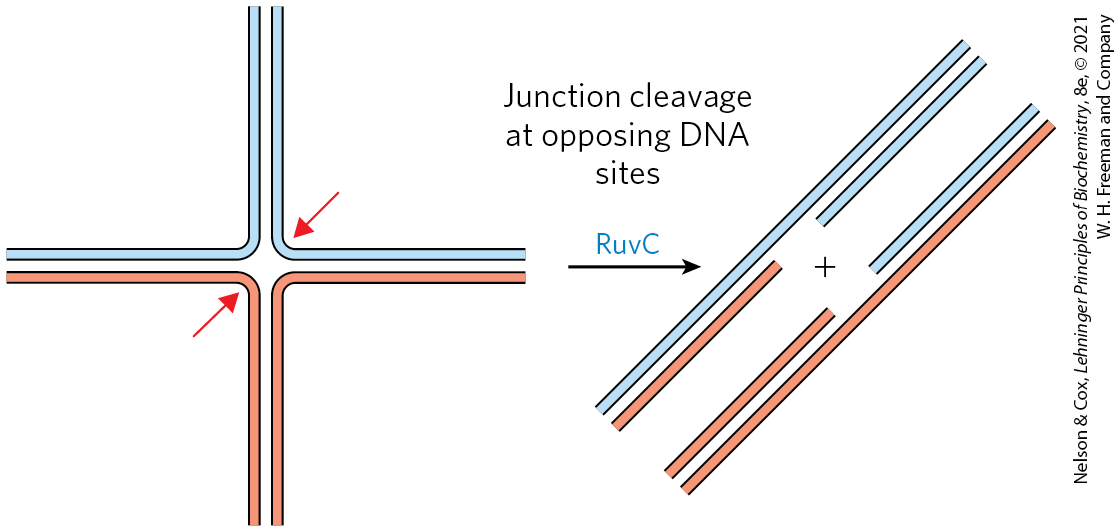
FIGURE 25-32 Resolution of a Holliday intermediate by the RuvC protein. RuvC is a specialized nuclease that binds to the RuvAB complex and cleaves the Holliday intermediate on opposing sides of the crossover junction (red arrows), so that two contiguous DNA arms remain in each product.
After the recombination steps are completed, the replication fork reassembles in a process called origin-independent restart of replication. Different combinations of four proteins (PriA, PriB, PriC, and DnaT) act with DnaC in several pathways to load DnaB helicase onto the reconstructed replication fork. The DnaG primase then synthesizes an RNA primer, and DNA polymerase III reassembles on DnaB to restart DNA synthesis. Complexes that include some combination of the PriA, PriB, PriC, and DnaT, along with DnaB, DnaC, and DnaG proteins, are called replication restart primosomes. In this way, the process of recombination is tightly intertwined with replication. One process of DNA metabolism supports the other.
Eukaryotic Homologous Recombination Is Required for Proper Chromosome Segregation during Meiosis
In eukaryotes, homologous genetic recombination has roles in replication and cell division, including the repair of stalled replication forks. Recombination occurs with the highest frequency during meiosis, the process by which diploid germ-line cells with two sets of chromosomes divide to produce haploid gametes (sperm cells or ova) in animals (haploid spores in plants) — each gamete having only one member of each chromosome pair (Fig. 25-33).
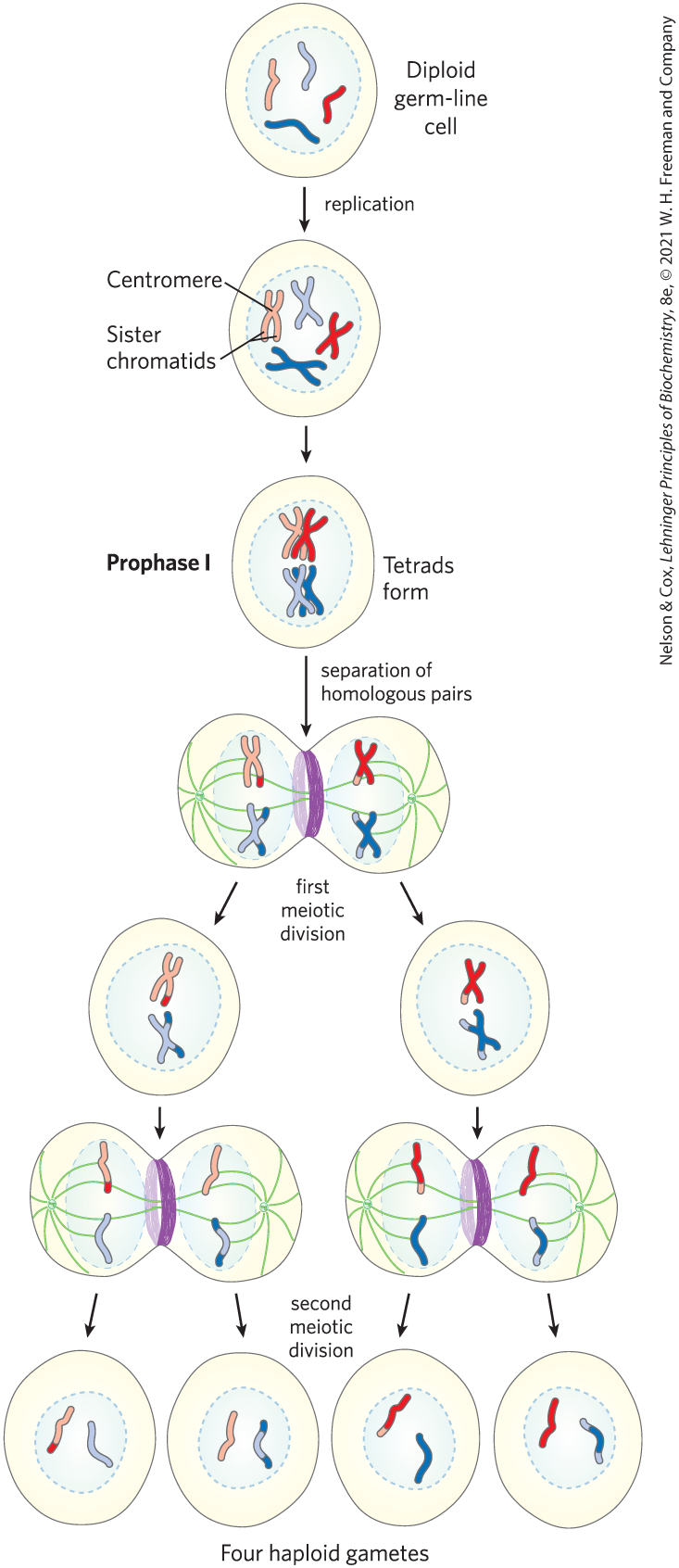
FIGURE 25-33 Meiosis in animal germ-line cells. The chromosomes of a hypothetical diploid germ-line cell (four chromosomes; two homologous pairs) replicate and are held together at their centromeres. Each replicated double-stranded DNA molecule is called a chromatid (sister chromatid). In prophase I, just before the first meiotic division, the two homologous sets of chromatids align to form tetrads, held together by covalent links at homologous junctions (chiasmata). Crossovers occur within the chiasmata (see Fig. 25-34). These transient associations between homologs ensure that the two tethered chromosomes segregate properly in the next step, when attached spindle fibers pull them toward opposite poles of the dividing cell in the first meiotic division. The products of this division are two daughter cells, each with two pairs of different sister chromatids. The pairs now line up across the equator of the cell in preparation for separation of the chromatids (now called chromosomes). The second meiotic division produces four haploid daughter cells that can serve as gametes. Each has two chromosomes, half the number of the diploid germ-line cell. The chromosomes have re-sorted and recombined.
Meiosis begins with replication of the DNA in the germ-line cell so that each DNA molecule is present in four copies. Each set of four homologous chromosomes (tetrad) exists as two pairs of sister chromatids, and the sister chromatids remain associated at their centromeres. The cell then goes through two rounds of cell division without an intervening round of DNA replication. In the first cell division, the two pairs of sister chromatids are segregated into daughter cells. In the second cell division, the two chromosomes in each sister chromatid pair are segregated into new daughter cells. In each division, the chromosomes to be segregated are drawn into the daughter cells by spindle fibers attached to opposite poles of the dividing cell. The two successive divisions reduce the DNA content to the haploid level in each gamete.
Proper chromosome segregation into daughter cells requires that physical links exist between the homologous chromosomes to be segregated. As the spindle fibers attach to the centromeres of chromosomes and start to pull, the links between homologous chromosomes create tension. This tension, sensed by cellular mechanisms not yet understood, signals that this pair of chromosomes or sister chromatids is properly aligned for segregation. Once the tension is sensed, the links are gradually dissolved and segregation proceeds. If improper spindle fiber attachment occurs (e.g., if the centromeres of a chromosome pair are attached to the same cellular pole), a cellular kinase senses the lack of tension and activates a system that removes the spindle attachments, allowing the cell to try again.
During the second meiotic division, the centromeric attachments between sister chromatids, augmented by cohesins deposited during replication (see Fig. 24-33), provide the physical links that are needed to guide segregation. However, during the first meiotic cell division, the two pairs of sister chromatids to be segregated are not related by a recent replication event and are not linked by cohesins or any other physical association. Instead, the homologous pairs of sister chromatids are aligned and new links are created by recombination, a process involving the breakage and rejoining of DNA (Fig. 25-34). This exchange, also referred to as crossing over, can be observed with the light microscope. Crossing over links the two pairs of sister chromatids together at points called chiasmata (singular, chiasma). Also during crossing over, genetic material is exchanged between the pairs of sister chromatids. These exchanges increase genetic diversity in the resulting gametes. The importance of meiotic recombination to proper chromosome segregation is well illustrated by the physiological and societal consequences of their failure (Box 25-2).
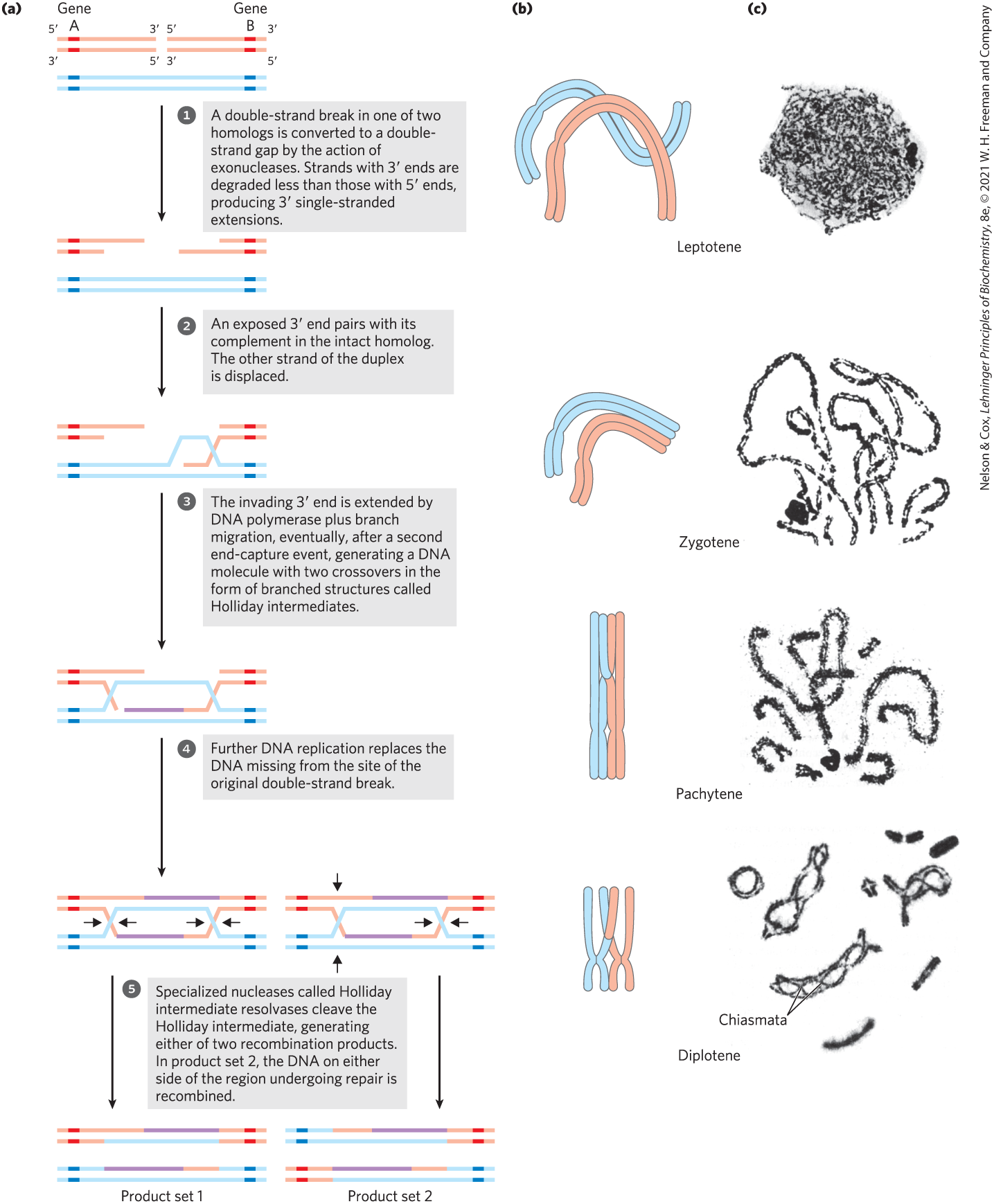
FIGURE 25-34 Recombination during prophase I in meiosis. (a) A model of double-strand break repair for homologous genetic recombination. The two homologous chromosomes (one shown in red, the other blue) involved in this recombination event have identical or very nearly identical sequences. Each of the two genes shown has different alleles on the two chromosomes. The steps are described in the text. (b) Crossing over occurs during prophase of meiosis I. The several stages of prophase I are aligned with the recombination processes shown in (a). Double-strand breaks are introduced and processed in the leptotene stage. The strand invasion and completion of crossover occur later. As homologous sequences in the two pairs of sister chromatids are aligned in the zygotene stage, synaptonemal complexes form and strand invasion occurs. The homologous chromosomes are tightly aligned by the pachytene stage. (c) Homologous chromosomes of a grasshopper, viewed at successive stages of meiotic prophase I. The chiasmata become visible in the diplotene stage. [(c) B. John, Meiosis, Figs 2.1a, 2.2a, 2.2b, 2.3a, Cambridge University Press, 1990. Reprinted with the permission of Cambridge University Press.]
A likely pathway for homologous recombination during meiosis is outlined in Figure 25-34a. The model has four key features. First, homologous chromosomes align. Second, a double-strand break is created in a DNA molecule, and the exposed ends are processed by an exonuclease, leaving a single-stranded extension with a free -hydroxyl group at the broken end (step ). Third, the exposed ends invade the intact duplex DNA of the homolog, and this is followed by branch migration and/or replication to create a pair of Holliday intermediates (steps to ). Fourth, cleavage of the two crossovers creates either of two pairs of complete recombinant products (step ). Notice the similarity of these steps to the bacterial recombinational repair processes outlined in Figure 25-29. The DNA strand invasion in eukaryotes is catalyzed by RecA-like recombinases called Rad51 and Dmc1. Loading of Rad51 onto DNA is promoted by Rad51 loading protein BRCA2 (analogous to the bacterial RecF, RecO, and RecR proteins).
In this double-strand break repair model for recombination, the ends are used to initiate the genetic exchange. Once paired with the complementary strand on the intact homolog, a region of hybrid DNA is created that contains complementary strands from two different parent DNAs (the product of step in Fig. 25-34a). Each of the ends can then act as a primer for DNA replication. Meiotic homologous recombination can vary in many details from one species to another, but most of the steps outlined above are generally present in some form. There are two ways to resolve the Holliday intermediate with a RuvC-like nuclease so that the two products carry genes in the same linear order as in the substrates — the original, unrecombined chromosomes (step ). If cleaved one way, the DNA flanking the region containing the hybrid DNA is not recombined; if cleaved the other way, the flanking DNA is recombined. Both outcomes are observed in vivo.
The homologous recombination illustrated in Figure 25-34 is an elaborate process that is essential to accurate chromosome segregation. Its molecular consequences for the generation of genetic diversity are subtle. To understand how this process contributes to diversity, we should keep in mind that the two homologous chromosomes that undergo recombination are not necessarily identical. The linear array of genes may be the same, but the base sequences in some of the genes may differ slightly (in different alleles). In a human, for example, one chromosome may contain the allele for hemoglobin A (normal hemoglobin) while the other contains the allele for hemoglobin S (the sickle cell mutation). The difference may consist of no more than one base pair among millions.
Crossing over is not an entirely random process, and “hot spots” have been identified on many eukaryotic chromosomes. However, the assumption that crossing over can occur with equal probability at almost any point along the length of two homologous chromosomes remains a reasonable approximation in many cases, and it is this assumption that permits the mapping of genes on a particular chromosome. The frequency of homologous recombination in any region separating two points on a chromosome is roughly proportional to the distance between the points, and this allows determination of the relative positions of different genes and the distances between those genes. The independent assortment of unlinked genes on different chromosomes (Fig. 25-35) makes another major contribution to the genetic diversity of gametes. These genetic realities guide many of the modern applications of genomics, such as defining haplotypes (see Fig. 9-26) or searching for disease genes in the human genome (see Fig. 9-30).
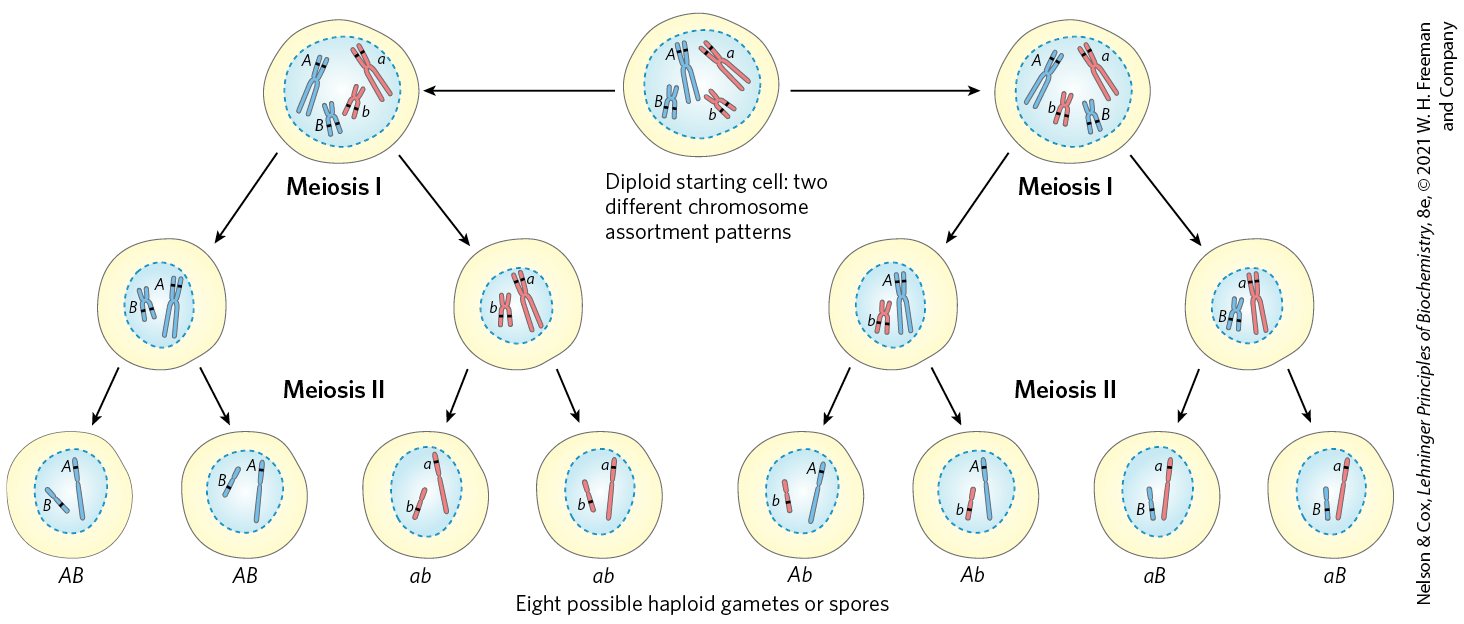
FIGURE 25-35 The contribution of independent assortment to genetic diversity. In this example, the two chromosomes have already been replicated to create two pairs of sister chromatids. Blue and red distinguish the sister chromatids of each pair. One gene on each chromosome is highlighted, with different alleles (A or a, B or b) in the homologs. Independent assortment can lead to gametes with any combination of the alleles present on the two different chromosomes. Crossing over (not shown here; see Fig. 25-34) would also contribute to genetic diversity in a typical meiotic sequence.
In the top center, a cell is shown that contains a nucleus with a large blue chromosome, a large red chromosome, a small blue chromosome, and a small red chromosome. Each chromosome has an “X” shape. Each chromosome has two black bands at the same location on each sister chromatid and these are labeled. The large blue chromosome has bands labeled upper case A, the large red chromosome has bands labeled lowercase a, the small blue chromosome has bands labeled lower case b, and the small red chromosome has bands labeled lower case b. Text below reads, Diploid starting cell: two different chromosome assortment patterns. Arrows point to the left and right to show two different paths through meiosis. The first arrow points left to a similar cell with two arrows extending down labeled meiosis Roman numeral 1. The left-hand arrow points to a cell containing a large blue chromosome labeled upper case A and a small blue chromosome containing uppercase B. The right-hand arrow points to a cell containing a large red chromosome containing lowercase a and a small red chromosome containing lowercase b. Two arrows labeled meiosis Roman numeral 2 point down from each of these cells. The left- and right-hand arrows from the left-hand cell both indicate cells labeled uppercase A uppercase B with one large blue chromosome labeled uppercase a and one small blue chromosome labeled uppercase B. The right-hand arrow points to a cell containing a large red chromosome labeled lowercase A and a small red chromosome containing lowercase a. The left- and right-hand arrows from the left-hand cell both indicate cells labeled lowercase a lowercase b with one large red chromosome labeled lowercase a and one small red chromosome labeled lowercase a. The second arrow from the cell at the top center points to a cell with two arrows extending down labeled meiosis Roman numeral 1. The left-hand arrow points to a cell containing a large blue chromosome labeled upper case A and a small blue chromosome containing uppercase B. The right-hand arrow points to a cell containing a large red chromosome containing lowercase a and a small red chromosome containing lowercase b. Two arrows labeled meiosis Roman numeral 2 point down from each of these cells. The left- and right-hand arrows from the left-hand cell both indicate cells labeled uppercase A uppercase B with one large blue chromosome labeled uppercase a and one small blue chromosome labeled uppercase B. The right-hand arrow points to a cell containing a large red chromosome labeled lowercase A and a small red chromosome containing lowercase a. The left- and right-hand arrows from the left-hand cell both indicate cells labeled lowercase a lowercase b with one large red chromosome labeled lowercase a and one small red chromosome labeled lowercase a. The second arrow from the cell at the top center points to a similar cell with two arrows extending down labeled meiosis Roman numeral 1. The left-hand arrow points to a cell containing a large blue chromosome containing uppercase a and a small red chromosome containing lowercase b. The right-hand arrow points to a cell containing a large red chromosome containing lowercase A and a small blue chromosome containing uppercase b. Two arrows labeled meiosis Roman numeral 2 point down from each of these cells. The left- and right-hand arrows from the left-hand cell both indicate cells labeled uppercase A lowercase b with one large blue chromosome labeled uppercase A and one small red chromosome labeled lowercase B. The left- and right-hand arrows from the right-hand cell both indicate cells labeled lowercase A uppercase b with one large red chromosome labeled lowercase A and one small blue chromosome labeled uppercase B. Text below reads, eight possible haploid gametes or spores.
As in bacteria, this recombination process is used to repair double-strand breaks that arise anywhere in the genome. In eukaryotes, these systems operate in the context of chromatin, rendering additional complexities to their regulation and damage detection mechanisms (Box 25-3). Homologous recombination thus serves at least three identifiable functions in eukaryotes: (1) it contributes to the repair of several types of DNA damage; (2) it provides, in eukaryotic cells, a transient physical link between chromatids that promotes the orderly segregation of chromosomes at the first meiotic cell division; and (3) it enhances genetic diversity in a population.
BOX 25-3 MEDICINE
How a DNA Strand Break Gets Attention
Each human chromosome contains many millions of DNA base pairs, all bound up in an elaborate chromatin structure (Chapter 24). If a strand break occurs somewhere in the DNA, how do the many proteins needed for its repair actually find it? The answer lies, at least in part, in a protein called poly-ADP ribose polymerase 1, or PARP1. PARP1 is a first responder, scanning the DNA for DNA damage and in particular for single-strand breaks. When it finds such sites, it binds and synthesizes an elaborate branched poly-ADP ribose polymer from an NAD precursor (Fig. 1). The polymers are attached to the PARP1 enzyme and also linked to some nearby proteins through Glu, Asp, or Lys residues. The resulting structure is a kind of signal, marking the chromosomal location of damage. A large number of DNA repair proteins bind to and are thus recruited to the poly-ADP ribose polymers, effecting DNA repair. If PARP1 activity is absent, repair is compromised and the number of single-strand breaks in all chromosomes increases. When the chromosome is replicated, the single-strand breaks become double-strand breaks (see Fig. 25-29).
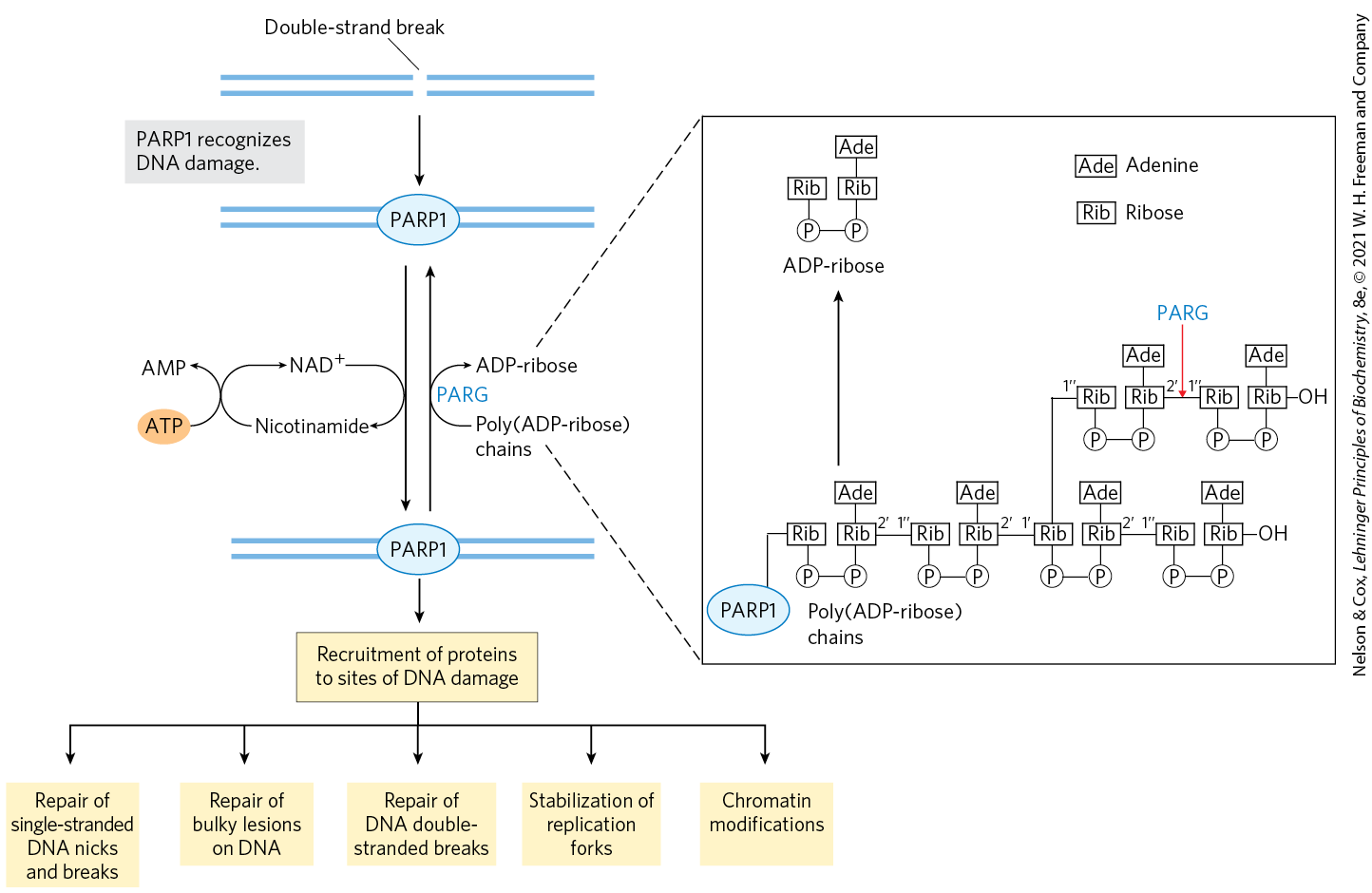
FIGURE 1 The activity and function of poly-ADP ribose polymerase in detecting DNA strand breaks and other types of damage. [Information from A. R. Chaudhuri and A. Nussenzweig, Nat. Rev. Mol. Cell Biol. 18:610, 2017, Fig. 1.]
Two blue horizontal strands of D N A are shown with a break near the center labeled double-strand break. An arrow points down next to a gray box that reads, P A R P 1 recognizes D N A damage. This yields a similar structure with an oval labeled P A R P 1 over the break. Upward- and downward-pointing arrows indicate a reversible reaction. The downward-arrow is met by a curved arrow showing the addition of N A D plus and loss of nicotinamide. A second arrow curves back from nicotinamide to N A D plus and is met by an arrow to the left showing that an orange oval labeled A T P is added and A M P is lost. The upward-pointing arrow is accompanied by a curved arrow showing the addition of poly (A D P-ribose) chains and loss of A D P-ribose with blue highlighted P A R G shown at the inflection point. This yields a double stranded D N A molecule with P A R P 1 at the place where there had been a break. An arrow points down to a yellow box reading, recruitment of proteins to sites of D N A damage. An arrow points down and breaks into five separate arrows pointing to five yellow boxes. From left to right, these boxes read: repair of single-stranded D N A nicks and breaks, repair of bulky lesions on D N A, repair of D N A double-stranded breaks, stabilization of replication forks, and chromatin modifications. Dashed lines from the reactions that convert poly (A D P-ribose) chains to A D P-ribose show a close-up of the process. A key indicates that a box labeled A d e represents adenine and a box labeled R i b represents ribose. Poly (A D P – ribose) chains is shown to the right of P A R P 1. A blue oval labeled P A R P 1 is bonded to R I b bonded to P below further bonded to P to the right bonded to R I b 2 prime above that is bonded to A d e above and to 1 prime prime R i b to the right bonded to P below bonded to P to the right bonded to R I b 2 prime above bonded to A d e above and to 1 prime R i b to the right that is bonded above and below. Below, it is bonded to P bonded to P bonded to R I b 2 prime above bonded to A d e above and to 1 prime prime R i b to the right bonded to P below bonded to P to the right bonded to R i b above bonded to A d e above and to O H to the right. Above, 1 prime R I b is bonded to 1 prime prime R I b bonded to P below bonded to P to the right bonded to R I b 2 prime above and bonded to A d e above across a bond indicated by an arrow from blue highlighted P A R G above to 1 prime prime R I b to the right bonded to P below bonded to P to the right bonded to R I b above boned to A d e above and to O H to the right. An arrow pointing up from the sequence immediately to the right of P A R P 1 is labeled A D P – ribose and shows R I b bonded to P below bonded to P to the right bonded to R I b above bonded to A d e above.
As we saw in Box 25-1, many malignant tumors have a defect in a DNA repair pathway. For example, breast or ovarian cancer is often associated with defects in double-strand break repair (e.g., in the genes encoding BRCA1 or BRCA2 or other proteins in the pathway). In these cells, the further loss of PARP1 activity is especially toxic, as single-strand breaks build up and chromosomes become broken during replication. This has led to the development of PARP1 inhibitors as a treatment for tumors in which double-strand break repair is defective. The first such pharmaceutical agent, olaparib, was approved for use in the United States in 2014. Many more PARP1 inhibitors have since been approved or are undergoing clinical trials. The effects have often been dramatic. For women with breast or ovarian tumors displaying deficiencies in BRCA1 or BRCA2 that have responded to more traditional therapies, subsequent maintenance treatment with PARP1 inhibitors has led to a fourfold increase in progression-free survival. PARP1 inhibitors are also showing promise for use with other breast and ovarian tumors, as well as other types of tumors, most of which have DNA repair deficiencies of some kind. As research continues, the use of PARP inhibitors is becoming an important part of the standard of care for a growing list of cancers.

Some Double-Strand Breaks Are Repaired by Nonhomologous End Joining
Double-strand breaks sometimes occur when recombinational DNA repair is not feasible, such as during phases of the cell cycle when no replication is occurring and no sister chromatids are present. At these times, another path is needed to avoid the cell death that would result from a broken chromosome. That alternative is provided by nonhomologous end joining (NHEJ). The broken chromosome ends are simply processed and ligated back together.
Nonhomologous end joining is an important pathway for double-strand break repair in all eukaryotes and has also been detected in some bacteria. The importance of NHEJ increases with genomic complexity, and the process accounts for most double-strand break repair outside meiosis in mammals. In yeast, most double-strand breaks are repaired by recombination, and only a few by NHEJ. NHEJ is a mutagenic process, and a smaller genome, such as that of yeast, has relatively little tolerance for the loss of information. The small genomic alterations may be tolerable in mammalian somatic cells, because they are balanced by the undamaged information on the homolog in each diploid cell, and in these non-germ-line cells the mutations are not inherited. In vertebrates, a loss of the genes encoding NHEJ function can produce a predisposition to cancer.
Unlike homologous recombinational repair, NHEJ does not conserve the original DNA sequence. The pathway in eukaryotes is illustrated in Figure 25-36. The reaction is initiated at the broken ends of a double-strand break by the binding of a heterodimer consisting of the proteins Ku70 and Ku80 (“KU” being the initials of the individual with scleroderma whose serum autoantibodies were used to identify this protein complex; the numbers refer to the approximate molecular weights of the subunits). The Ku proteins are conserved in almost all eukaryotes and act as a kind of molecular scaffold to assemble the other protein components. Ku70-Ku80 interacts with another protein complex containing a protein kinase called DNA-PKcs and a nuclease known as Artemis. Once the complex is assembled, the two broken DNA ends are synapsed (held together). DNA-PKcs autophosphorylates in several locations and also phosphorylates Artemis. Artemis, when phosphorylated, acquires an endonuclease function that can remove or single-stranded extensions or hairpins that might be present at the ends. The DNA ends are then separated with the aid of a helicase, and strands from the two different ends are annealed at locations where short regions of complementarity are encountered. Artemis cleaves any unpaired DNA segments that are created. Small DNA gaps are filled by a DNA polymerase, Pol or Pol . Finally, the nicks are sealed by a protein complex consisting of XRCC4 (x-ray cross complementation group), XLF (XRCC4-like factor), and DNA ligase IV.
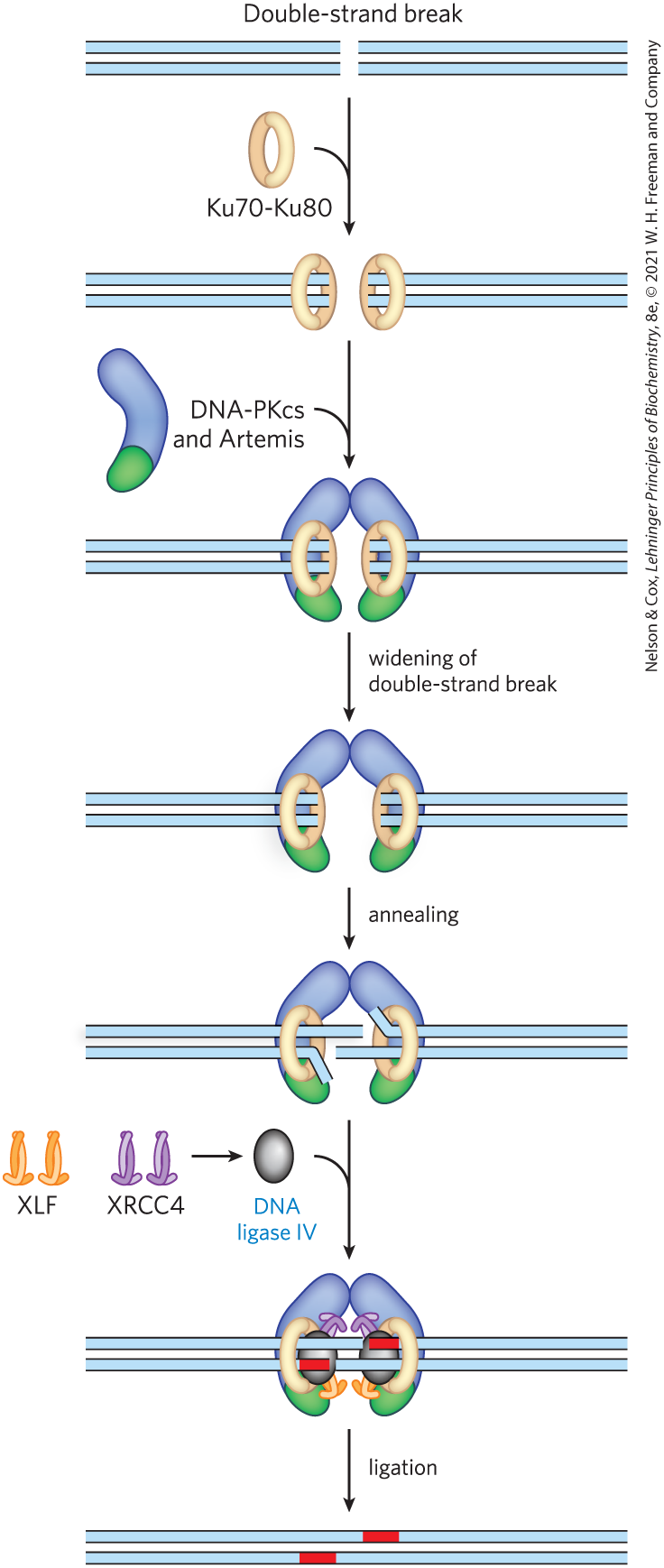
FIGURE 25-36 Nonhomologous end joining. The Ku70-Ku80 complex is the first to bind the DNA ends, followed by a complex including DNA-PKcs and the nuclease Artemis. These proteins then recruit a complex consisting of XRCC4, XLF, and DNA ligase IV. Either of two DNA polymerases, Pol or Pol (not shown), subsequently extends the annealed DNA strands, as needed, before ligation. [Information from J. M. Sekiguchi and D. O. Ferguson, Cell 124:260, 2006, Fig. 1.]
A blue horizontal double-stranded piece of D N A has a break in the center labeled double-strand break. An arrow points down accompanied by a curved line showing the addition of a ring-shaped structure darker on the back half than on the front half and labeled K u 70 – K u 80. This yields a similar piece of broken D N A with a ring on each of the broken ends. The left-hand piece has a ring on its right side and the right-hand piece has a ring on its right side. An arrow points down accompanied by a curved line showing the addition of D N A – P K c s and Artemis. This is a purple comma-shaped structure with a green tip. This yields a similar product in which there is a purple comma-shaped structure behind each ring with the green portion behind the bottom of the ring. These purple structures are bent so that the top halves come together above the D N A. An arrow points down accompanied by text reading, widening of double-strand break. This shows a similar figure in which the purple structures have bent and the sides of the D N A molecules have moved farther apart. An arrow points down labeled annealing. This yields a product in which the D N A strands have moved into the opening. The top left blue strand has moved through the ring almost halfway across the opening. The top right blue strand has moved through the ring and bent upward. The lower right blue strand has extended over halfway across the opening, so that it extends beneath the upper left blue strand above. An arrow points down accompanied by a curved line showing the addition of a gray oval labeled D N A ligase Roman numeral 4. X L F and X R C C 4 are shown to the left with an arrow pointing to blue highlighted D N A ligase Roman numeral 4. X K F is shown as a light orange strand and a dark orange strand twisted together so that the ends extend out to left and right and there is a long, somewhat oval piece above. X R C C 4 is similar but purple. This yields a structure in which gray ovals are next to the openings in the circles around the broken D N A with X R C C R present at the top of each circle and X L F present at the bottom of each circle. A red piece of D N A extends left from the end of the upper right-hand piece and a similar red piece extends right from the end of the lower left-hand piece. An arrow labeled ligation points down to show that this produces a blue double stranded D N A molecule with a small red piece in each strand. The red piece in the top strand is slightly to the right of the red piece in the bottom strand.
DNA ends are not joined randomly by NHEJ. Instead, when a double-strand break occurs, the ends are generally constrained by the structure of chromatin and thus remain close together. Very rare events linking end sequences that are normally far apart in the chromosome, or are on different chromosomes, may be responsible for occasional dramatic and usually deleterious genomic rearrangements.
Site-Specific Recombination Results in Precise DNA Rearrangements
Homologous genetic recombination can involve any two homologous sequences. The second general type of recombination, site-specific recombination, is a very different type of process: recombination is limited to specific sequences. Recombination reactions of this type occur in virtually every cell, filling specialized roles that vary greatly from one species to another. Examples include regulation of the expression of certain genes and promotion of programmed DNA rearrangements in embryonic development or in the replication cycles of some viral and plasmid DNAs. Each site-specific recombination system consists of an enzyme called a recombinase and a short (20 to 200 bp), unique DNA sequence where the recombinase acts (the recombination site). One or more auxiliary proteins may regulate the timing or outcome of the reaction.
There are two general classes of site-specific recombination systems, which rely on either Tyr or Ser residues in the active site. In vitro studies of many site-specific recombination systems in the tyrosine class have elucidated some general principles, including the fundamental reaction pathway (Fig. 25-37a). Several of these enzymes have been crystallized, revealing structural details of the reaction. A separate recombinase recognizes and binds to each of two recombination sites on two different DNA molecules or within the same DNA. One DNA strand in each site is cleaved at a specific point within the site, and the recombinase becomes covalently linked to the DNA at the cleavage site through a phosphotyrosine bond (step ). The transient protein-DNA linkage preserves the phosphodiester bond that is lost in cleaving the DNA, so high-energy cofactors such as ATP are unnecessary in subsequent steps. The cleaved DNA strands are rejoined to new partners to form a Holliday intermediate, with new phosphodiester bonds created at the expense of the protein-DNA linkage (step ). An isomerization then occurs (step ), and the process is repeated at a second point within each of the two recombination sites (steps and ). In systems that employ an active-site Ser residue, both strands of each recombination site are cut concurrently and rejoined to new partners without the Holliday intermediate. In both types of systems, the exchange is always reciprocal and precise, regenerating the recombination sites when the reaction is complete. We can view a recombinase as a site-specific endonuclease and ligase in one package.
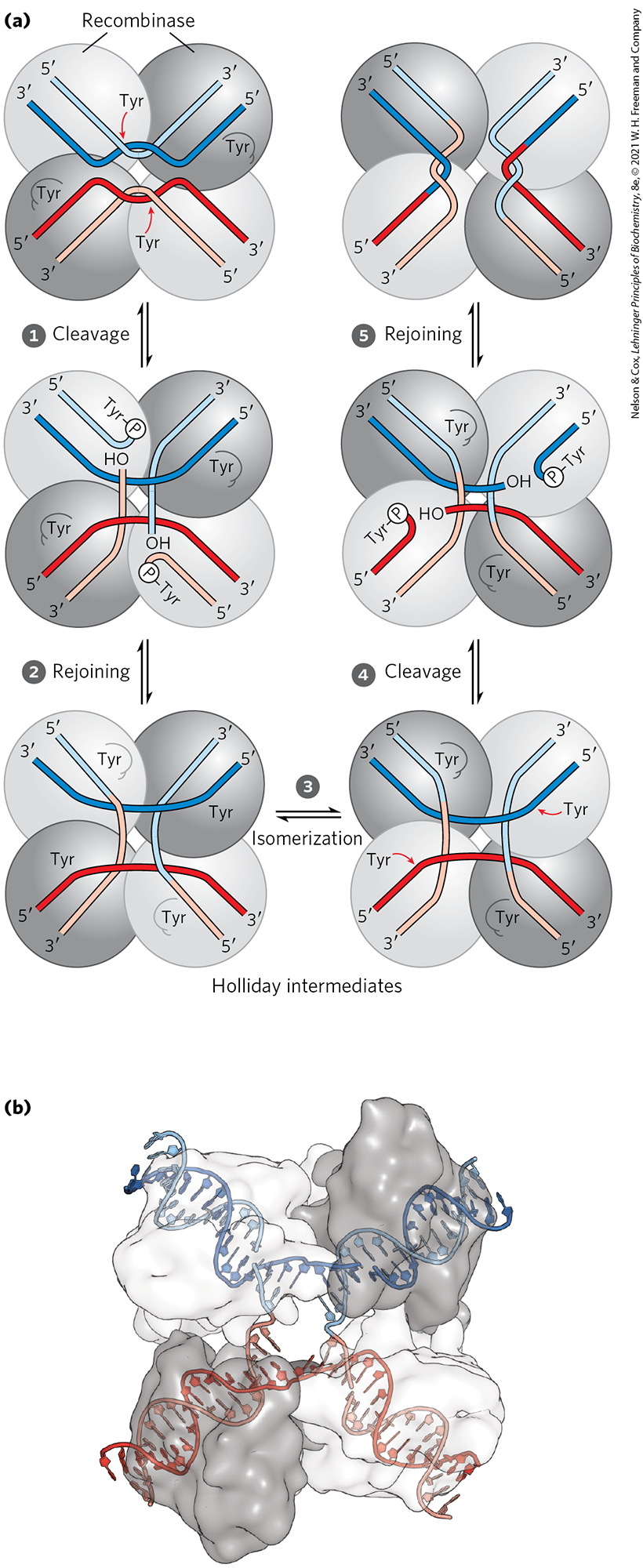
FIGURE 25-37 A site-specific recombination reaction. (a) The reaction shown here is for a common class of site-specific recombinases called integrase-class recombinases (named after bacteriophage integrase, the first recombinase characterized). These enzymes use Tyr residues as nucleophiles at the active site. The reaction is carried out within a tetramer of identical subunits. Recombinase subunits bind to a specific sequence, the recombination site. Two dimeric complexes, each bound to a single site in the DNA, come together to form the tetrameric complex shown here. One strand in each DNA is cleaved at particular points in the sequence. The nucleophile is the group of an active-site Tyr residue, and the product of rejoining is a covalent phosphotyrosine link between protein and DNA. After isomerization , the cleaved strands join to new partners, producing a Holliday intermediate. Steps and complete the reaction by a process similar to the first two steps. The original sequence of the recombination site is regenerated after recombining the DNA flanking the site. These steps occur within a complex of multiple recombinase subunits that sometimes includes other proteins not shown here. (b) Surface contour model of a four-subunit integrase-class recombinase called the FLP recombinase, bound to a Holliday intermediate (shown with light blue and dark blue helix strands). The protein has been rendered transparent so that the bound DNA is visible. Another group of recombinases, called the resolvase/invertase family, use a Ser residue as nucleophile at the active site. [(b) Data from PDB ID 1P4E, P. A. Rice and Y. Chen, J. Biol. Chem. 278:24,800, 2003.]
Part a shows four spheres in a roughly cuboidal shape with lighter spheres at the upper left and lower right and darker spheres at the lower left and upper right. These spheres are labeled recombinase. A light blue strand begins at its 5 prime end at the upper left of the upper left sphere and runs diagonally down to the place where the two top spheres meet, then bends upward to end at its 3 prime end. A dark blue strand begins at its 3 prime end below and runs beneath the light blue strand into the very top of the lower left sphere, then loops over the bottom bend of the light blue sphere down again, and then up to end at its 5 prime end beneath the 3 prime end of the light blue sphere. A red arrow points from T y r to the place where the blue strand crosses behind the light blue strand. T y r is also shown at the lower right of the upper right sphere. The bottom two spheres show the same pattern flipped so that the strands run up instead of down and with dark and light red strands instead of blue strands. Step 1: Cleavage. Upward- and downward-pointing arrows indicate a reversible reaction. This yields a product in which the blue strand follows a similar trajectory, but the light blue sphere begins at its 5 prime end at the upper right and runs diagonally down to curve up to a white circle labeled P bonded to T y r at the right side of the upper left sphere. In the upper right sphere, the light blue strand runs from its 3 prime end at the upper right down to the lower left, then vertically down to O H in the lower right sphere. The same pattern is present with the red lines except that they are flipped so that they run in opposite directions to the blue strands. Step 2: Rejoining. Upward- and downward-pointing arrows labeled rejoining indicate a reversible reaction. The dark blue and dark red strands remain the same. The light blue strand beginning at 5 prime at the upper left runs down to the lower right of the upper left sphere, then joins the light red strand and bends back to end at its 3 prime end to the lower left of the lower left sphere. The 3 prime end of the light blue sphere at the upper right of the upper right sphere runs to the lower left and then bends down and then to the lower right. Its lower right portion is light red. This structure forms a central square around a small opening between the spheres with light red on the left, dark blue on the top, light blue on the right, and dark red below. In this illustration, the upper left sphere is labeled (a). Step 3: Isomerization. Right- and left-pointing arrows labeled isomerization indicate a reversible reaction. This illustration resembles the previous illustration except that the dark gray spheres are now at the upper left and lower right and the light gray spheres are at the lower left and upper right. The stands are in the same orientations despite the movement of the spheres. Text beneath these spheres and those in the previous step reads, Holliday intermediates. T y r is shown with a red arrow pointing to the dark red strand near where it meets the light red strand in the lower left sphere and pointing to the dark blue strand near where it meets the light blue strand in the upper right sphere. Step 4: Cleavage. Upward- and downward-pointing arrows indicate a reversible reaction. This yields a similar structure in which the central opening has become smaller, the red strand ends in O H right after crossing the light red strand, and the dark blue strand ends at O H just after crossing the light blue strand. The remaining dark red piece still begins at 5 prime at the lower left, but runs up and then curves left to end at a white circle labeled P bonded to T y r. The remaining dark blue piece still starts at 5 prime at the upper right and runs to the lower left, then bends to bind to a white circle labeled P bonded to T y r. Step 5: Rejoining. Upward- and downward-pointing arrows indicate a reversible reaction. On the left, the 5 prime end of the light blue strand runs to the lower right to meet the light red strand, which bends left and then back right before looping back to end at the 3 prime end at the lower left. The dark blue 3 prime end is below the 5 prime end of the light blue piece and run right across the light red piece where it loops to the left before looping back under the light red piece and joining a bright red piece to end at the 5 prime end to the lower right. The 3 prime end of the light blue piece at the upper right runs to the lower left, then loops right, then back left, then back right to join a light red piece to end at the 5 prime end at the lower right. The 5 prime end of the dark blue piece begins beneath is and runs left before becoming red, then runs over the light blue piece before looping back under it to run diagonally to end at the 3 prime end at the lower right. Part b shows a surface contour illustration with roughly spherical white pieces at the upper left and lower right and roughly spherical gray pieces to the lower left and upper right. A blue piece begins at the upper left of the upper white piece, runs down to just above the opening between the pieces where it is open, and then runs up to end at the upper right in the dark gray piece. A red piece of D N A begins at the lower left, runs up to the central open area where it connects with the blue strands above, then bends down to the lower right.
The sequences of the recombination sites recognized by site-specific recombinases are partially asymmetric (nonpalindromic), and the two recombining sites align in the same orientation during the recombinase reaction. The outcome depends on the location and orientation of the recombination sites (Fig. 25-38). If the two sites are on the same DNA molecule, the reaction either inverts or deletes the intervening DNA, determined by whether the recombination sites have the opposite or the same orientation, respectively. If the sites are on different DNAs, the recombination is intermolecular; if one or both DNAs are circular, the result is an insertion. Some recombinase systems are highly specific for one of these reaction types and act only on sites with particular orientations.
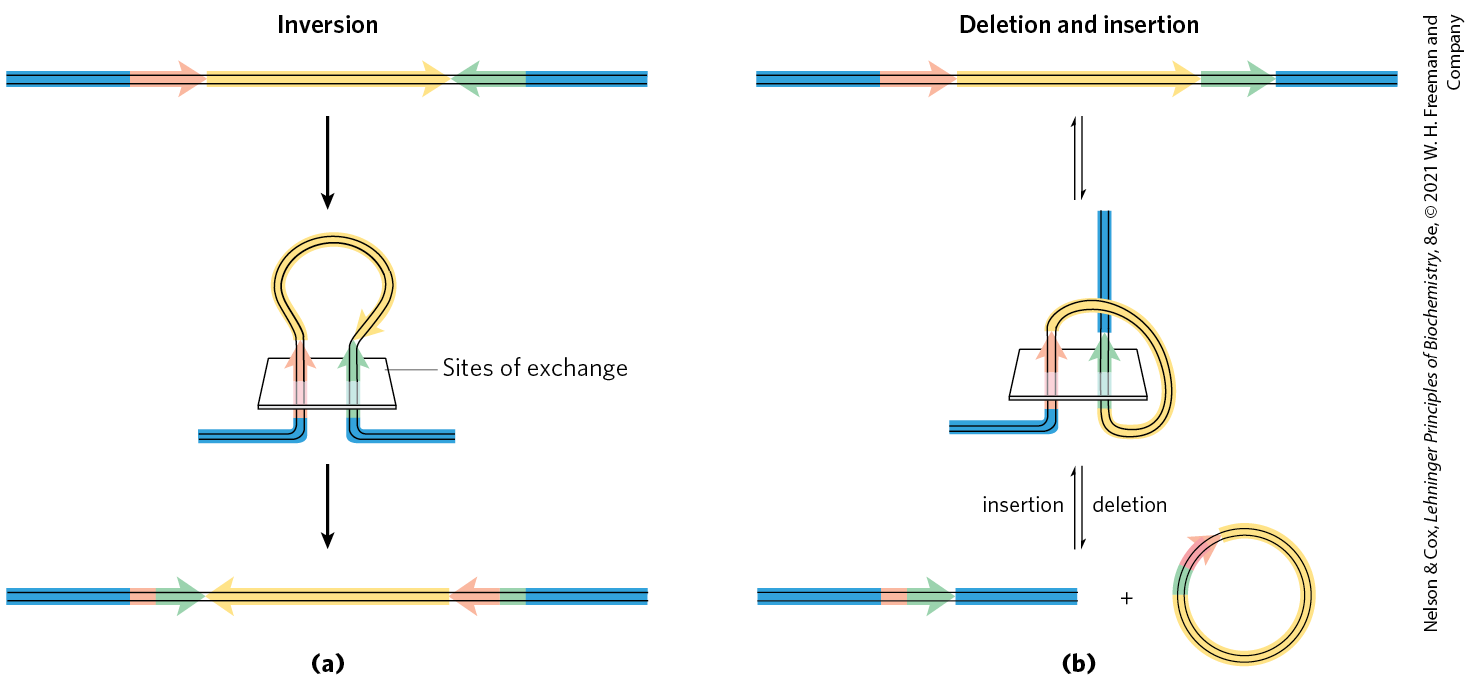
FIGURE 25-38 Effects of site-specific recombination. The outcome of site-specific recombination depends on the location and orientation of the recombination sites (red and green) in a double-stranded DNA molecule. Orientation here (shown by arrowheads) refers to the order of nucleotides in the recombination site, not the direction. (a) Recombination sites with opposite orientation in the same DNA molecule. The result is an inversion. (b) Recombination sites with the same orientation, either on one DNA molecule, producing a deletion, or on two DNA molecules, producing an insertion.
Part a shows a horizontal strand labeled inversion. It is blue on the left, then has a short red arrow pointing right, then has a long yellow arrow pointing right, then has a short green arrow pointing left, then has a blue piece. An arrow points down to show the same strand bent into a loop that passes through a plane. The blue pieces run horizontally beneath the plane. The left-hand piece bends up into a light red arrow that points up through the plane to reach a yellow loop. The right-hand piece bends up into a green arrow that points up through the plane to reach the other end of the same yellow loop. The plane is labeled, sites of exchange. An arrow points down to show that this yields a strand with blue, then a small re piece, than a green arrow pointing right, then a yellow arrow pointing left, then a short red arrow pointing left, then a short green piece, then a blue piece. Part a shows a horizontal strand labeled deletion and insertion. It is blue on the left, then has a short red arrow pointing right, then has a long yellow arrow pointing right, then has a short green arrow pointing right, then has a blue piece. Upward- and downward-pointing arrows indicate a reversible reaction. A blue piece runs horizontally, then bends to a red arrow pointing up through a plane to a yellow loop that bends right and runs beneath the plane before reaching a green arrow that points back through the plane to a vertical blue piece. Upward- and downward-pointing arrows indicate a reversible reaction The downward-pointing arrow is labeled deletion and the upward-pointing arrow is labeled insertion. This yields a blue piece attached to a short red piece attached to a green arrow pointing right attached to a blue piece plus a yellow circle with a small green piece at the upper left joined to a small red arrow pointing clockwise.
Complete chromosomal replication can require site-specific recombination. Recombinational DNA repair of a circular bacterial chromosome, while essential, sometimes generates deleterious byproducts. The resolution of a Holliday intermediate at a replication fork by a nuclease such as RuvC, followed by completion of replication, can give rise to one of two products: the usual two monomeric chromosomes or a contiguous dimeric chromosome (Fig. 25-39). In the latter case, the covalently linked chromosomes cannot be segregated to daughter cells at cell division, and the dividing cells become “stuck.” A specialized site-specific recombination system in E. coli, the XerCD system, converts the dimeric chromosomes to monomeric chromosomes so that cell division can proceed. The reaction is a site-specific deletion (Fig. 25-38b). This is another example of the close coordination between DNA recombination processes and other aspects of DNA metabolism.
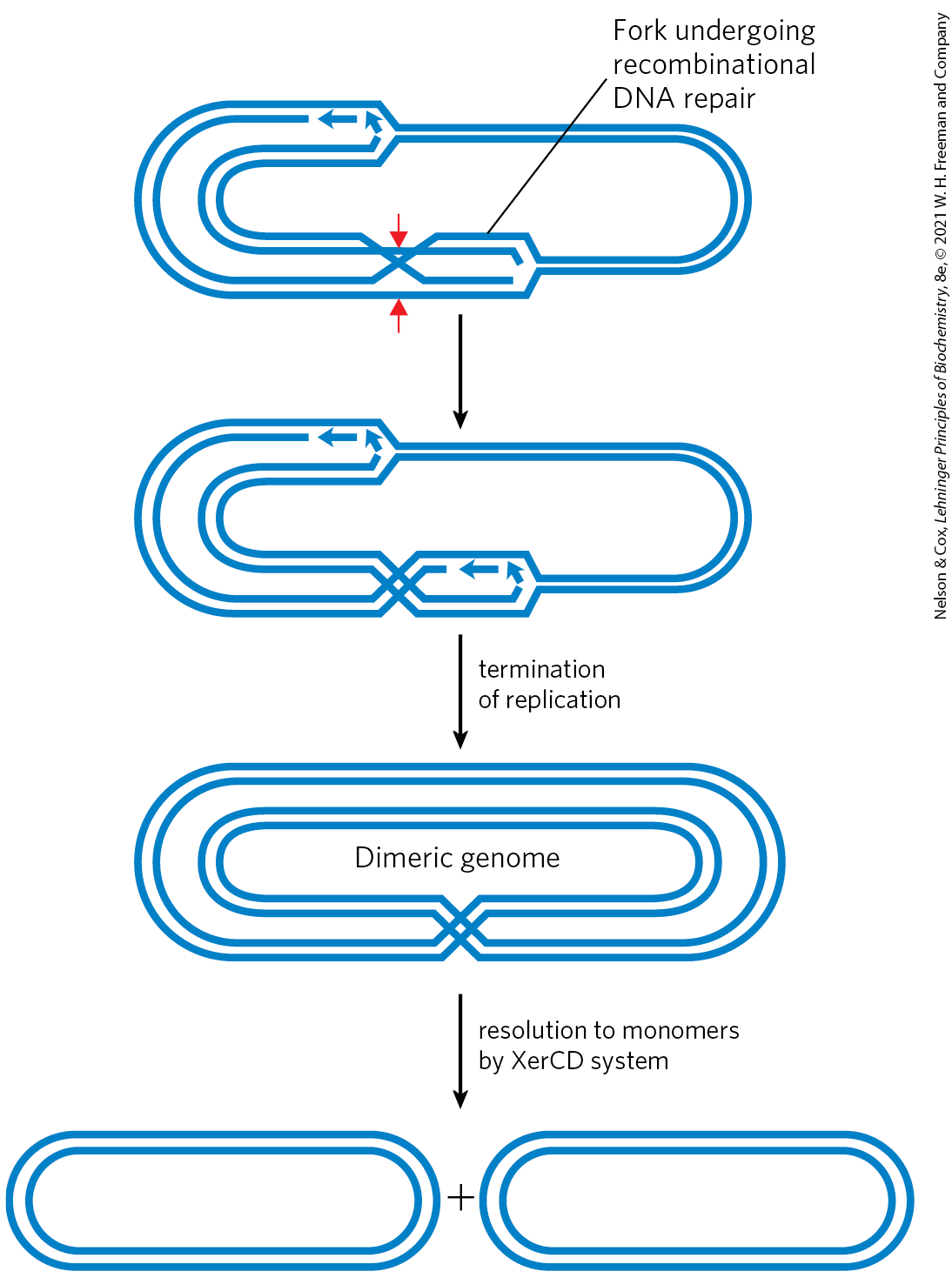
FIGURE 25-39 DNA deletion to undo a deleterious effect of recombinational DNA repair. The resolution of a Holliday intermediate during recombinational DNA repair (if cut at the points indicated by the red arrows) can generate a contiguous dimeric chromosome. A specialized site-specific recombinase in E. coli, XerCD, converts the dimer to monomers, allowing chromosome segregation and cell division to proceed.
A double-stranded piece of D N A forms an oval at the right with forks at the upper left and near the bottom center. The upper left fork has two small arrows pointing from the fork to te left, but most of the strands lining the fork are continuous. The fork at the bottom center has a top strand that bends to the lower left to cross a strand from the upper right, creating an “X” shape with red arrows pointing to it from above and below. Text reads, fork undergoing recombinational D N A repair. An arrow points down to show that the open region tot eh right of the “X” now has Okazaki fragments across the top and a continuous stand across the bottom. An arrow labeled termination of replication points downward. This yields a dimeric genome, shown as a double stranded outer oval and a double stranded inner oval with an “X” at the bottom center connecting them. An arrow points down accompanied by text reading, resolution to monomers by X e r C D system. This yields two separate double stranded ovals.
Transposable Genetic Elements Move from One Location to Another
We now consider the third general type of recombination system: recombination that allows the movement of transposable elements, or transposons. These segments of DNA, found in virtually all cells, move, or “jump,” from one place on a chromosome (the donor site) to another on the same or a different chromosome (the target site). DNA sequence homology is not usually required for this movement, called transposition; the new location is determined more or less randomly. Insertion of a transposon in an essential gene could kill the cell, so transposition is tightly regulated and usually very infrequent. Transposons are perhaps the simplest of molecular parasites, adapted to replicate passively within the chromosomes of host cells. In some cases they carry genes that are useful to the host cell, and thus exist in a kind of symbiosis with the host.
Bacteria have two classes of transposons. Insertion sequences (simple transposons) contain only the sequences required for transposition and the genes for the proteins (transposases) that promote the process. Complex transposons contain one or more genes in addition to those needed for transposition. These extra genes might, for example, confer resistance to antibiotics and thus enhance the survival chances of the host cell. The spread of antibiotic-resistance elements among disease-causing bacterial populations that is rendering some antibiotics ineffectual (p. 887) is mediated to a large degree by transposition.
Bacterial transposons vary in structure, but most have short repeated sequences at each end that serve as binding sites for the transposase. When transposition occurs, a short sequence at the target site (5 to 10 bp) is duplicated to form an additional short repeated sequence that flanks each end of the inserted transposon (Fig. 25-40). These duplicated segments result from the cutting mechanism used to insert a transposon into the DNA at a new location.
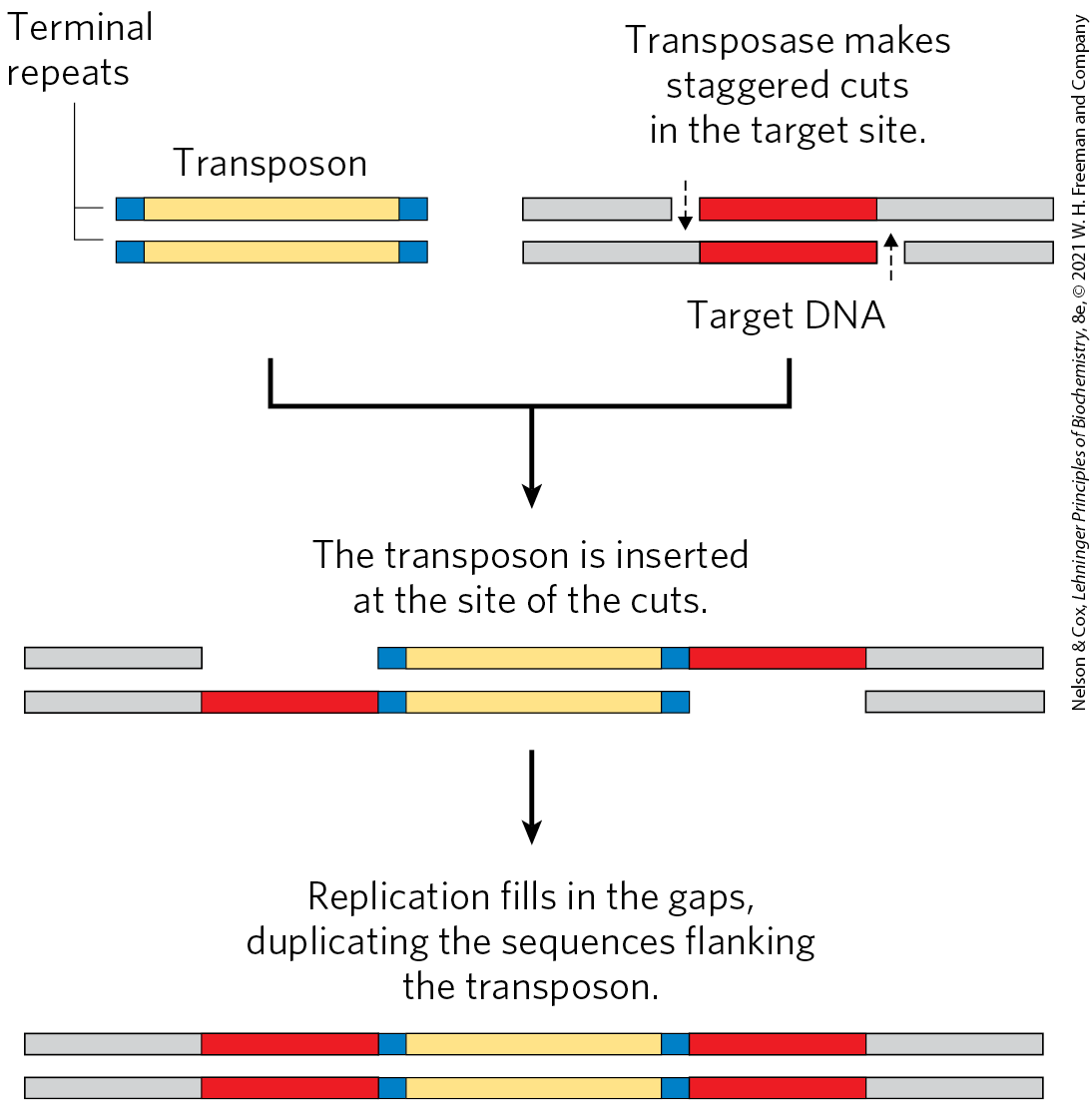
FIGURE 25-40 Duplication of the DNA sequence at a target site when a transposon is inserted. The sequences duplicated following transposon insertion are shown in red. These sequences are generally only a few base pairs long, so their size relative to that of a typical transposon is greatly exaggerated in this drawing.
Two horizontal bars at the upper left are labeled transposon. They are yellow in the center with small blue ends labeled terminal repeats. To the right, target D N A is shown as two bars that are gray to the left and right with red regions in the center. Text above reads, transposase makes staggered cuts in the target site. A dashed arrow points down along the left side of the top red bar and a similar dashed arrow points up along the right side of the bottom red bar. Text below the red bars reads, target D N A. An arrow points down from both structures, the transposon and the target D N A. Text above reads, the transposon is inserted at the site of the cuts. The top strand has a gray piece, then a break, then a transposon, then a red piece, then a gray piece. The bottom piece has a gray piece, then a red piece beneath the space above, then a transposon, then a space beneath the red piece above, then a gray piece. An arrow points down. Text reads, replication fills in the gaps, duplicating the sequences flanking the transposon. This results in a similar structure in which the open regions are filled by red sequences.
There are two general pathways for transposition in bacteria. In direct (or simple) transposition (Fig. 25-41, left), cuts on each side of the transposon excise it, and the transposon moves to a new location. This leaves a double-strand break in the donor DNA that must be repaired. At the target site, a staggered cut is made (as in Fig. 25-40), the transposon is inserted into the break, and DNA replication fills in the gaps to duplicate the target-site sequence. In replicative transposition (Fig. 25-41, right), the entire transposon is replicated, leaving a copy behind at the donor location. A cointegrate is an intermediate in this process, consisting of the donor region covalently linked to DNA at the target site. Two complete copies of the transposon are present in the cointegrate, both having the same relative orientation in the DNA. In some well-characterized transposons, the cointegrate intermediate is converted to products by site-specific recombination, in which specialized recombinases promote the required deletion reaction.
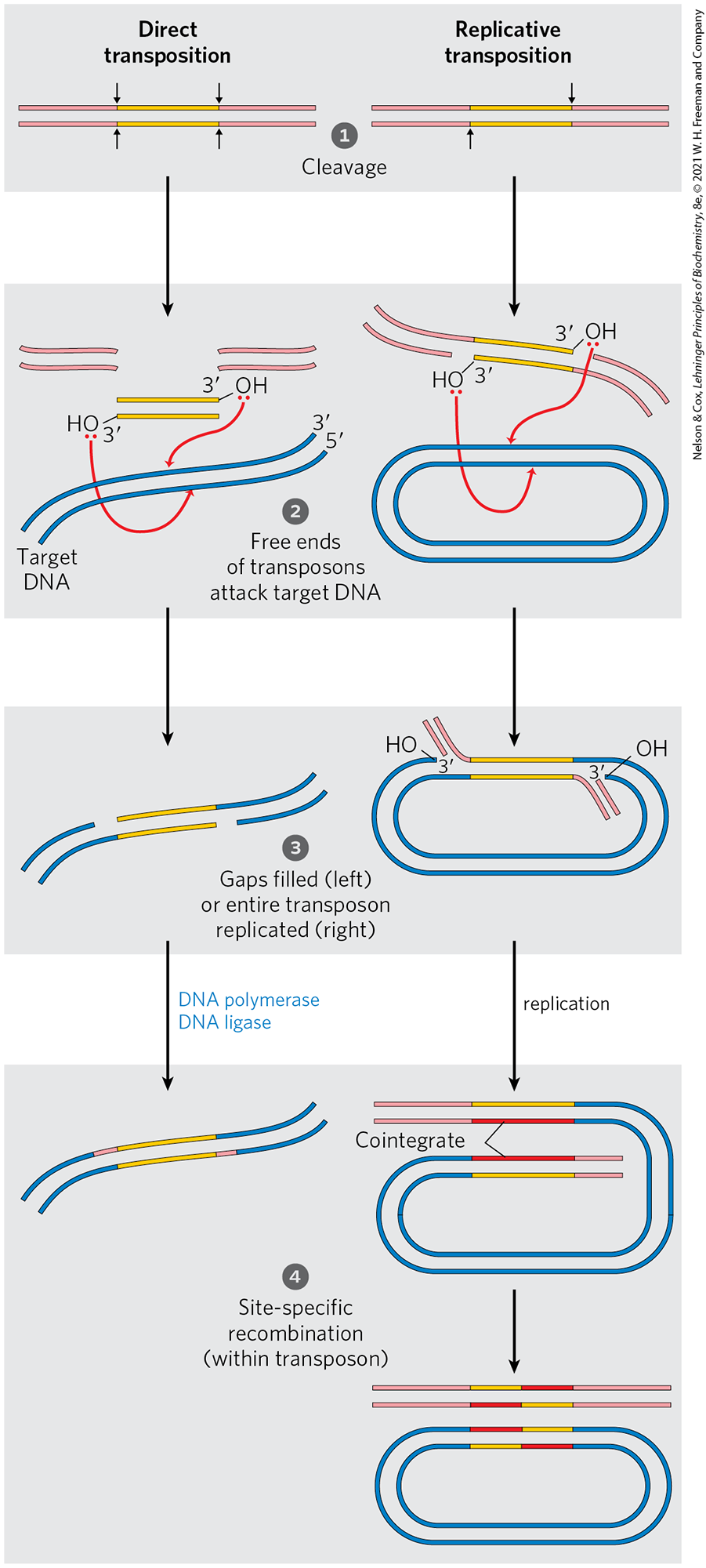
FIGURE 25-41 Two general pathways for transposition: direct (simple) and replicative. The DNA is first cleaved on each side of the transposon, at the sites indicated by arrows. The liberated -hydroxyl groups at the ends of the transposon act as nucleophiles in a direct attack on phosphodiester bonds in the target DNA. The target phosphodiester bonds are staggered (not directly across from each other) in the two DNA strands. The transposon is now linked to the target DNA. In direct transposition (left), replication fills in gaps at each end to complete the process. In replicative transposition (right), the entire transposon is replicated to create a cointegrate intermediate. The cointegrate is often resolved later, with the aid of a separate site-specific recombination system. The cleaved host DNA left behind after direct transposition is either repaired by DNA end joining or degraded (not shown); the latter outcome can be lethal to the organism.
Step 1: Cleavage. Two double stranded molecules are shown. Each has red pieces to the sides and yellow pieces in the center. The left-hand piece has arrows pointing down on either side of the top yellow piece and up at either side of the bottom yellow piece. This is labeled direct transposition. The right-hand piece is similar but has fewer arrows. It has an arrow pointing up to the left side of the bottom yellow piece and an arrow pointing down to the right side of the upper yellow piece. This is labeled replicative transposition. Arrows point down from each structure. Step 2: Free ends of transposons attack target D N A. In direct transposition, two double stranded red pieces are shown, a double stranded yellow piece is shown with the top right ending with 3 prime O H and the lower left ending with 3 prime O H. Each O has a red pair of electrons. Blue double stranded target D N A is below. Red arrows point from each red pair of electron on O to a location on the target D N A, one on the top piece and one on the bottom piece. In replicative transposition, a strand is shown with red attached to a yellow piece ending with 3 prime O H, then there is a loose red piece. Below, there is a loose red piece, then the end of a yellow piece with 3 prime O H that has a red piece bonded to the other end of the yellow piece. Each O has a red pair of electrons. An oval of double stranded D N A is below. Red arrows point from the red pairs of electrons on O to the target D N A, one pointing to the inner strand and one pointing to the outer strand. Arrows point down from each side. Step 3: Gaps filled (left) or entire transposon replicated (right). On the left, a blue piece has a small space before a yellow piece joined with a blue piece. Beneath, a blue piece is joined to a yellow piece and then there is a space before there is another blue piece. On the right, two yellow pieces are in the double-stranded oval with one in the outer strand and one in the outer strand. The yellow piece in the outer strand ends with a red piece that bends up with a parallel short red piece running up to its left with 3 prime below next to O H on the end of the adjacent blue piece to its left. The yellow piece in the inner strand ends with a red piece that bends inward. A small red piece runs along it with 3 prime above next to O H at the end of the blue piece. An arrow points down on the left accompanied by text reading, blue highlighted D N A polymerase, D N A ligase. This yields two strands. The top strand has a blue piece, then a short red piece, then a yellow piece, then a blue piece. The bottom strand has a blue piece, then a yellow piece, then a short red piece, then a blue piece. Step 4: Site-specific recombination (within transposon). The oval double-stranded D N A has separated so that there is a bottom piece that is almost an oval with a top piece that has a top strand that is red then light red with a lower strand that is yellow and then light red. The blue piece coils around below and back across the top so that its lower strand has red above the red below that joins to light red to the left. The strand above has yellow above the red in the strand below and this piece is also joined to light red. Lines joining the red pieces read, cointegrate. An arrow points down to show a blue oval with the top center having red followed by yellow in the top strand and yellow followed by red in the bottom strand. Above this, there are two linear strands. The top strand is light red, then yellow, then red, then light red. The bottom strand is light red, then red, then yellow, then light red.
Eukaryotes also have transposons, structurally similar to bacterial transposons, and some use similar transposition mechanisms. In other cases, however, the mechanism of transposition seems to involve an RNA intermediate. Evolution of these transposons is intertwined with the evolution of certain classes of RNA viruses. Both are described in the next chapter. As illustrated in Figure 9-25, nearly half of the human genome is made up of various types of transposable elements.
Immunoglobulin Genes Assemble by Recombination
Some DNA rearrangements are a programmed part of development in eukaryotic organisms. An important example is the generation of complete immunoglobulin genes from separate gene segments in vertebrate genomes. A human (like other mammals) is capable of producing millions of different immunoglobulins (antibodies) with distinct binding specificities, even though the human genome contains only ~20,000 genes. Recombination allows an organism to produce an extraordinary diversity of antibodies from a limited DNA-coding capacity. Studies of the recombination mechanism reveal a close relationship to DNA transposition and suggest that this system for generating antibody diversity may have evolved from an ancient cellular invasion by transposons.
We can use the human genes that encode proteins of the immunoglobulin G (IgG) class to illustrate how antibody diversity is generated. Immunoglobulins consist of two heavy and two light polypeptide chains (see Fig. 5-20). Each chain has two regions: a variable region, with a sequence that differs greatly from one immunoglobulin to another, and a region that is virtually constant within a class of immunoglobulins. There are also two distinct families of light chains, kappa and lambda, which differ somewhat in the sequences of their constant regions. For all three types of polypeptide chains (heavy chain, and kappa and lambda light chains), diversity in the variable regions is generated by a similar mechanism. The genes for these polypeptides are divided into segments, and the genome contains clusters with multiple versions of each segment. The joining of one version of each gene segment creates a complete gene.
Figure 25-42 depicts the organization of the DNA encoding the kappa light chains of human IgG and shows how a mature kappa light chain is generated. In undifferentiated cells, the coding information for this polypeptide chain is separated into three segments. The V (variable) segment encodes the first 95 amino acid residues of the variable region, the J (joining) segment encodes the remaining 12 residues of the variable region, and the C segment encodes the constant region. The genome contains 40 different V segments, 5 different J segments, and 1 C segment.
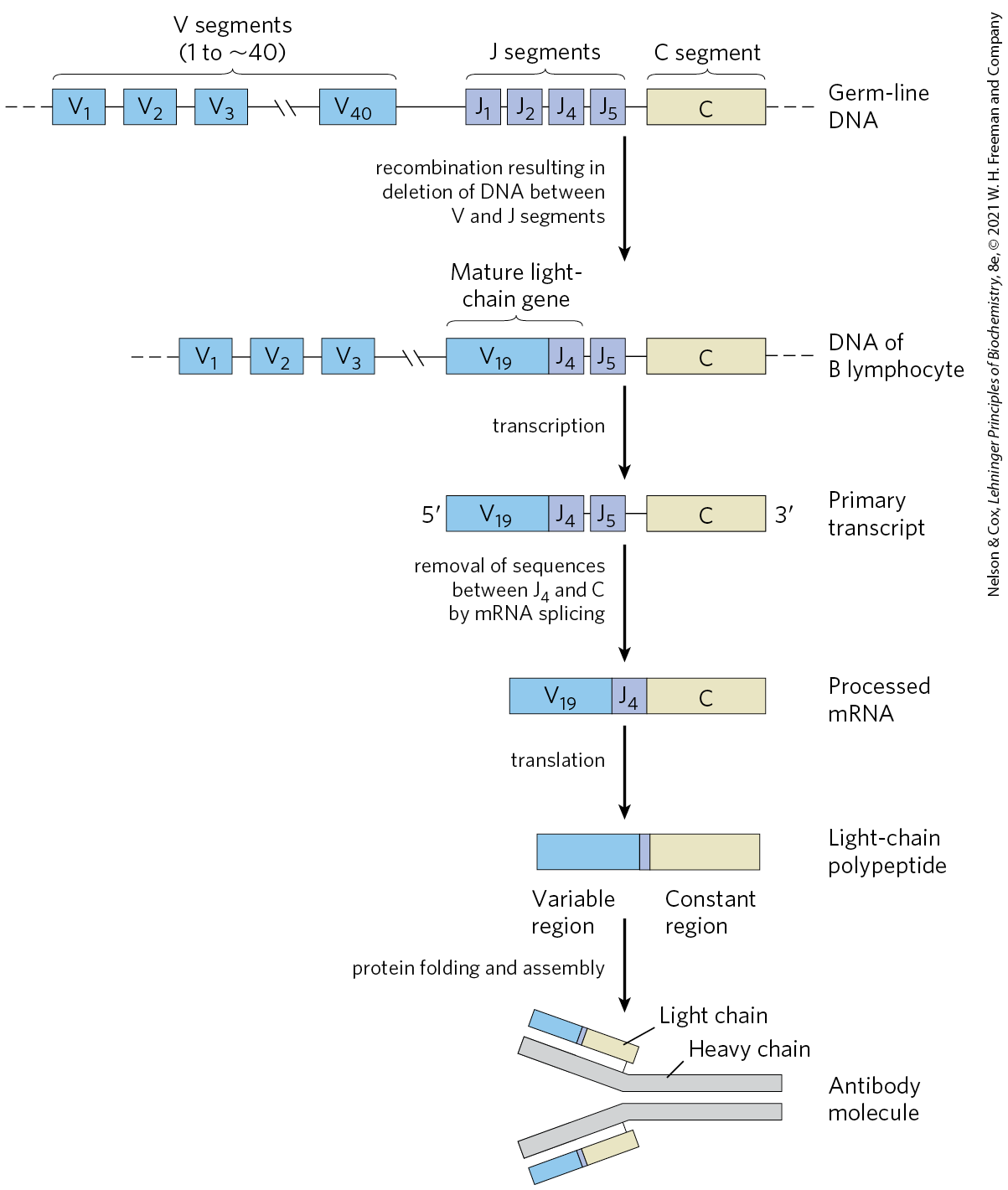
FIGURE 25-42 Recombination of the V and J gene segments of the human IgG kappa light chain. At the top is shown the arrangement of IgG-coding sequences in a stem cell of the bone marrow. Recombination deletes the DNA between a particular V segment and a J segment. Transcription and RNA splicing, as described in Chapter 26, produces the light-chain polypeptide. The light chain can combine with any of 5,000 possible heavy chains to produce an antibody molecule.
A chain across the top is labeled germ-line D N A. From right to left, it has an olive rectangle labeled C representing the C segment joined to four purple boxes labeled J segments that are J 5, J 4, J 2, and J 1 from right to left, bonded to a long chain of V segments (1 to approximately 40) shown in blue boxes as V 40, then a break, then V 3, V 2, and V 1 before dashes to the left. An arrow points down accompanied by text that reads, recombination resulting in deletion of D N A between V and J segments. This results in a similar structure labeled D N A of B lymphocyte that begins with an olive rectangle labeled C joined to a purple box labeled J 5 joined to a mature light-chain gene consisting of a purple box labeled J 4 joined to a blue box labeled V 19, then a break, then blue boxes labeled V 3, the V 2, then V 1 followed by three dashes. An arrow points down labeled transcription. This yields a primary transcript. It has 3 prime to the right of an olive box labeled C joined to a purple box labeled J 5 joined to J 4 adjacent to a blue box labeled V 19 that has a left end labeled 5 prime. An arrow labeled translation points down to a light-chain polypeptide, shown as an olive constant region connected by a narrow purple band to a blue variable region. An arrow points down labeled protein folding and assembly. This yields an antibody molecule. This has two gray chains labeled heavy chain that are parallel at the right and then branch t the upper and lower left. Where the heavy chain bends upward, it is connected to an olive box next to a narrow purple box next to a blue box. This is labeled light chain. A similar structure is bound to the bottom half of the heavy chain where the two strands separate.
As a stem cell in the bone marrow differentiates to form a mature B lymphocyte, one V segment and one J segment are brought together by a specialized recombination system (Fig. 25-42). During this programmed DNA deletion, the intervening DNA is discarded. There are about possible V–J combinations. The recombination process is not as precise as the site-specific recombination described earlier, so additional variation occurs in the sequence at the V–J junction. This increases the overall variation by a factor of at least 2.5, so the cells can generate about different V–J combinations. The final joining of the V–J combination to the C region is accomplished by an RNA-splicing reaction after transcription, a process described in Chapter 26.
The recombination mechanism for joining the V and J segments is illustrated in Figure 25-43. Just beyond each V segment and just before each J segment lie recombination signal sequences (RSSs). These are bound by proteins called RAG1 and RAG2 (products of the recombination activating gene). The RAG proteins catalyze the formation of a double-strand break between the signal sequences and the V (or J) segments to be joined. The V and J segments are then joined with the aid of a second complex of proteins.
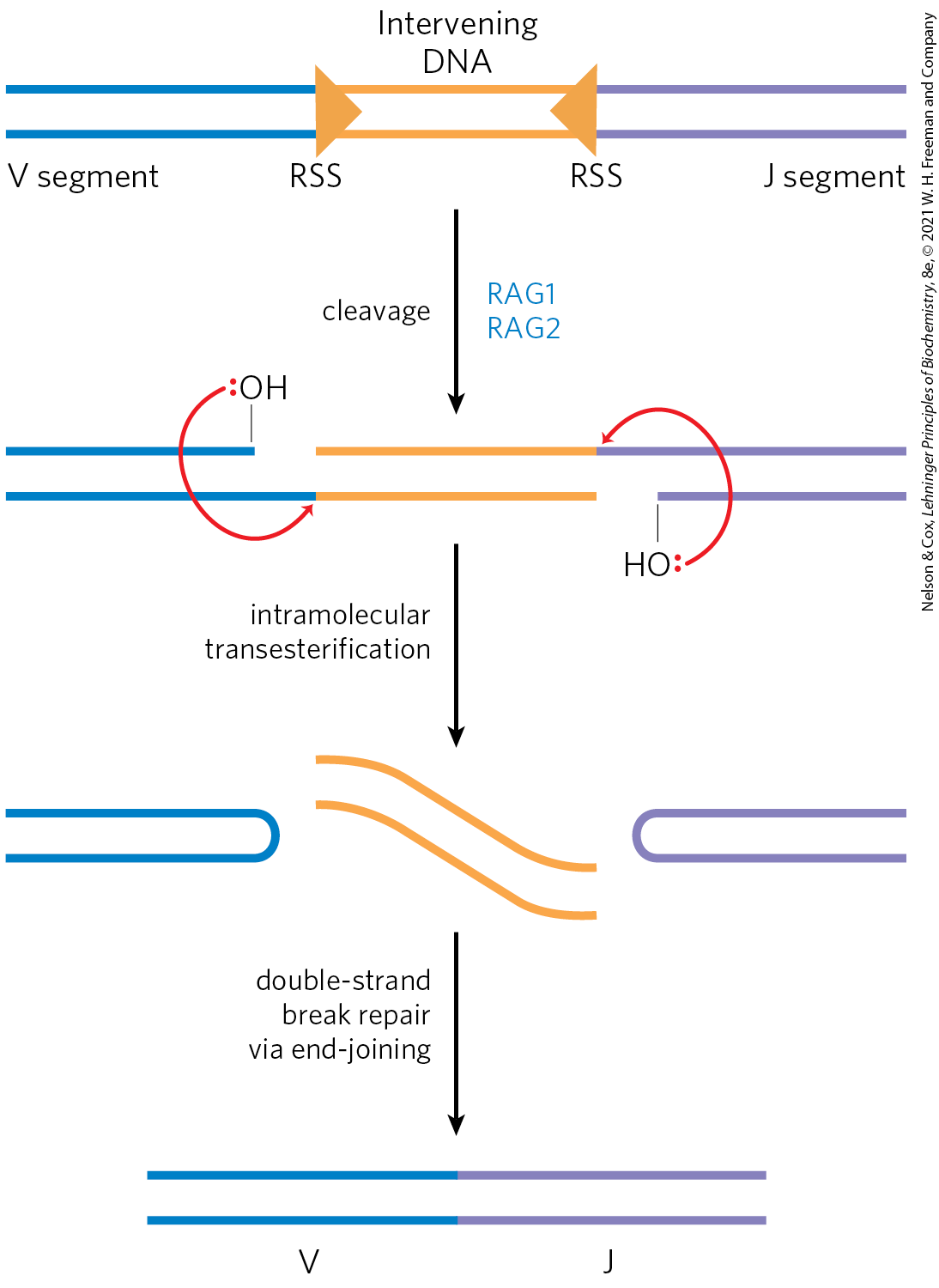
FIGURE 25-43 Mechanism of immunoglobulin gene rearrangement. The RAG1 and RAG2 proteins bind to the recombination signal sequences (RSSs) and cleave one DNA strand between the RSS and the V (or J) segments to be joined. The liberated hydroxyl then acts as a nucleophile, attacking a phosphodiester bond in the other strand to create a double-strand break. The resulting hairpin bends on the V and J segments are cleaved, and the ends are covalently linked by a complex of proteins specialized for end-joining repair of double-strand breaks.
A double-stranded piece of D N A is blue on the left, orange in the middle, and purple on the right. The blue region is labeled V segment. An orange triangle pointing right labeled R S S marks the beginning of the orange region, which is labeled intervening D N A and ends with a triangle pointing left. The purple region is labeled J segment. An arrow points down labeled blue highlighted R A G 1, R A G 2 and cleavage. This yields a similar molecule in which the top blue piece has had its right end removed and now ends with a bond to O H above with a red pair of electrons on O. The lower left end of the purple segment is missing and has been replaced by a bond to O H with a red pair of electrons on O. The orange triangles are gone. Red arrows point from the red electrons on the left-hand O to the place that the bottom blue piece and orange piece come together and from the red electrons on the right-hand O to the place where the right end of the upper orange piece and upper purple piece come together. An arrow points down labeled intramolecular transesterification. This yields a blue piece on the left that has a top piece that curves around and loops back to the left, two wavy yellow lines, and a purple piece that runs horizontally to the left, curves down, and runs back to the right. An arrow points down labeled double-strand break repair via end-joining This yields a double-stranded molecule with a left-hand blue piece labeled V joined to a purple piece to the right labeled J.
The genes for the heavy chains and the lambda light chains form by similar processes. Heavy chains have more gene segments than light chains, with more than 5,000 possible combinations. Because any heavy chain can combine with any light chain to generate an immunoglobulin, each human has at least possible IgGs. And additional diversity is generated by high mutation rates (of unknown mechanism) in the V sequences during B-lymphocyte differentiation. Each mature B lymphocyte produces only one type of antibody, but the range of antibodies produced by the B lymphocytes of an individual organism is clearly enormous.
Did the immune system evolve in part from ancient transposons? The mechanism for generation of the double-strand breaks by RAG1 and RAG2 mirrors several reaction steps in transposition (Fig. 25-43). In addition, the deleted DNA, with its terminal RSSs, has a sequence structure found in most transposons. In the test tube, RAG1 and RAG2 can associate with this deleted DNA and insert it, transposonlike, into other DNA molecules (probably a rare reaction in B lymphocytes). Although we cannot know for certain, the properties of the immunoglobulin gene rearrangement system suggest an intriguing origin in which the distinction between host and parasite has become blurred by evolution.
SUMMARY 25.3 DNA Recombination
- DNA sequences are rearranged in recombination reactions, usually in processes tightly coordinated with DNA replication or repair.
- Homologous genetic recombination can take place between any two DNA molecules that share sequence homology. In bacteria, recombination serves mainly as a DNA repair process, focused on reactivating stalled or collapsed replication forks or on the general repair of double-strand breaks.
- In eukaryotes, recombination is essential to ensure accurate chromosome segregation during the first meiotic cell division. It also helps to create genetic diversity in the resulting gametes.
- Nonhomologous end joining provides an alternative mechanism for the repair of double-strand breaks, especially in eukaryotic cells.
- Site-specific recombination occurs only at specific target sequences, and this process can also involve a Holliday intermediate. Recombinases cleave the DNA at specific points and ligate the strands to new partners. This type of recombination is found in virtually all cells, and its many functions include DNA integration and regulation of gene expression.
- In almost all cells, transposons use recombination to move within or between chromosomes.
- In vertebrates, a programmed recombination reaction related to transposition joins immunoglobulin gene segments to form immunoglobulin genes during B-lymphocyte differentiation.
 In bacteria, homologous genetic recombination is primarily a DNA repair process, and in this context (as noted in
In bacteria, homologous genetic recombination is primarily a DNA repair process, and in this context (as noted in 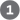 The -ending strand at the break is degraded to create a single-stranded extension, which is then used in
The -ending strand at the break is degraded to create a single-stranded extension, which is then used in  a strand invasion process, pairing the invading single strand with its complementary strand within the adjacent duplex.
a strand invasion process, pairing the invading single strand with its complementary strand within the adjacent duplex.  Migration of the branch (shown in the box) can create a Holliday intermediate.
Migration of the branch (shown in the box) can create a Holliday intermediate.  Cleavage of the Holliday intermediate by specialized nucleases, followed by ligation, restores a viable replication fork. The replisome is reloaded onto this structure (not shown), and replication continues. Arrowheads represent ends.
Cleavage of the Holliday intermediate by specialized nucleases, followed by ligation, restores a viable replication fork. The replisome is reloaded onto this structure (not shown), and replication continues. Arrowheads represent ends. ). Notice the similarity of these steps to the bacterial recombinational repair processes outlined in
). Notice the similarity of these steps to the bacterial recombinational repair processes outlined in  Homologous recombination thus serves at least three identifiable functions in eukaryotes: (1) it contributes to the repair of several types of DNA damage; (2) it provides, in eukaryotic cells, a transient physical link between chromatids that promotes the orderly segregation of chromosomes at the first meiotic cell division; and (3) it enhances genetic diversity in a population.
Homologous recombination thus serves at least three identifiable functions in eukaryotes: (1) it contributes to the repair of several types of DNA damage; (2) it provides, in eukaryotic cells, a transient physical link between chromatids that promotes the orderly segregation of chromosomes at the first meiotic cell division; and (3) it enhances genetic diversity in a population. MEDICINE
MEDICINE
 DNA sequences are rearranged in recombination reactions, usually in processes tightly coordinated with DNA replication or repair.
DNA sequences are rearranged in recombination reactions, usually in processes tightly coordinated with DNA replication or repair.