Project-Specific JSON Control Files¶
The CoNNECT custom developed support tools utilize JSON control files from each project’s CODE bids directory (Fig. 13). The files are named as <project_identifier>_<data_type>_<process>_input.json. Data types are specified in Table 4 and available processes are flirt and bet.
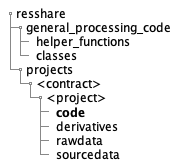
Fig. 13 Directory tree showing the location of a Project’s JSON control files.¶
Data Type |
Source Image Description |
|---|---|
struc |
T1-weighted image |
asl |
arterial spin labeling or cerebral blood flow image |
apt |
amide proton transfer-weighted source or MTRasym image |
flair |
T2 FLAIR image |
T2 |
T2 or T2* image |
Brain Extraction JSON Control File¶
Brain extraction parameters (Table 5) for FSL BET or ANTs Brain Extraction can be found in “<project_identifier>_<input-datatype>_bet_input.json”.
See also
This file contains the inputs described in the nipype python extension manual for FSL BET or ANTs Brain Extraction.
Sample Brain Extraction JSON control files
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
|
OPTIONAL |
string |
free text to provide a brief description |
type |
REQUIRED |
string |
brain extraction program: FSL or ANTs |
bet_params |
REQUIRED |
dictionary |
brain extraction parameters as described in Table 6 and Table 7 |
BET-Specific parameters¶
FSL BET¶
Key Name |
Required Level |
Data Type |
Description |
|---|---|---|---|
args |
OPTIONAL |
string |
Additional parameters to the command |
center |
OPTIONAL |
list |
3 integer items specifying the center of gravity in voxels |
environ |
OPTIONAL |
dictionary |
Environment variables |
frac |
OPTIONAL |
float |
Fractional intensity threshold |
functional |
OPTIONAL |
boolean |
NOT SUPPORTED |
ignore_exception |
OPTIONAL |
boolean |
print an error instead of throwing an exception in case the interface fails to run |
mask |
OPTIONAL |
boolean |
create binary mask image |
mesh |
OPTIONAL |
boolean |
generate a vtk mesh brain surface |
no_output |
OPTIONAL |
boolean |
don’t generate segmented output |
outline |
OPTIONAL |
boolean |
create surface outline image |
output_type |
OPTIONAL |
string |
FSL output type (‘NIFTI_PAIR’, ‘NIFTI_PARI_GZ’, ‘NIFTI_GZ’, ‘NIFTI’) |
padding |
OPTIONAL |
boolean |
improve BET if FOV is very small in Z |
radius |
OPTIONAL |
integer |
Head radius |
reduce_bias |
OPTIONAL |
boolean |
Bias field and neck cleanup |
remove_eyes |
OPTIONAL |
boolean |
eye & optic nerve cleanup (can be useful in SIENA) |
robust |
OPTIONAL |
boolean |
Robust brain centre estimation (iterates BET several times) |
skull |
OPTIONAL |
boolean |
Creates a skull image |
surfaces |
OPTIONAL |
boolean |
run bet2 and then betsurf to get additional skull and scalp surfaces (includes registrations) |
t2_guided |
OPTIONAL |
boolean |
Requires a dictionary titled T2 as described in Table 8 as with creating surfaces, when also feeding in non-brain-extracted T2 (includes registrations) |
terminal_output |
OPTIONAL |
string |
Control terminal output (stream, allatonce, file, none) |
threshold |
OPTIONAL |
boolean |
apply thresholding to segmented brain image and mask |
vertical gradient |
OPTIONAL |
float |
Vertical gradient in fractional intensity threshold (-1, 1) |
ANTs Brain Extraction¶
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
brain_probability_template |
REQUIRED |
string |
full path to an existing brain probability mask |
brain_template |
REQUIRED |
string |
full path to an anatomical template |
args |
OPTIONAL |
string |
additional parameters to the command |
debug |
OPTIONAL |
boolean |
if True, runs a faster version of the script. Only for testing. Implies -u 0 |
dimension |
OPTIONAL |
integer |
image dimension (2 or 3) |
environ |
OPTIONAL |
dictionary |
Environment variables |
extraction_registration_mask |
OPTIONAL |
string |
full path to a mask (in template space) used during registration for brain extraction |
image_suffix |
OPTIONAL |
string |
Any of standard ITK formats, nii.gz is default |
keep_temporary_files |
OPTIONAL |
integer |
Keep brain extraction/segmentation warps, etc (default = 0) |
Optional Parameters¶
T2/T2 FLAIR
Note
T2 functionality has not been implemented or evaluated.
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
input_bids_location |
REQUIRED |
string |
Location of original, non-brain extracted T2 or T2 FLAIR image: ‘rawdata’ or ‘derivatives’ |
input_bids_parameters |
REQUIRED |
dictionary |
A bids filename dictionary as explained in NEEDS REFERENCE |
FLIRT JSON Control File¶
FLIRT parameters can be found in “<project_identifier>_<input-datatype>_flirt_input.json” (see Table 4 for a list of available input data types). The available parameters are provided in Table 9.
See also
flirt.py also supports the execution of brain extraction, controlled through its own JSON file
Sample FLIRT JSON control files
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
|
OPTIONAL |
string |
free text to provide a brief description |
fslroi |
OPTIONAL |
boolean |
perform fslroi to extract a single volume from input 4D image. Extracts the center volume unless the ‘volume’ key of the main_image_params |
bet |
OPTIONAL |
boolean |
run brain extraction |
inclusion_list |
OPTIONAL |
list[string] |
list of strings for option search inclusion criteria |
exclusion_list |
OPTIONAL |
list[string] |
list of strings for option search exclusion criteria |
main_image_params |
OPTIONAL |
dictionary |
parameters as described in Table 10 |
reference_image_params |
OPTIONAL |
dictionary |
parameters as described in Table 11 |
flirt_params |
REQUIRED |
dictionary |
FLIRT parameters as described in Our FLIRT input Table |
secondary_image_params |
OPTIONAL |
dictionary |
parameters as described in Table 12 |
standard_reference_params |
OPTIONAL |
dictionary |
parameters as described in Table 13 |
Required Input Image Parameters¶
main_image_params
These keys are used to identify the main input image for registration.
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
input_bids_labels |
REQUIRED |
dictionary |
A bids filename dictionary as explained in NEEDS REFERENCE |
output_bids_location |
REQUIRED |
string |
bids derivatives sub-folder (derivatives -> sub-XXX -> ses-YYY - > output_bids_location) |
output_matrix_base |
REQUIRED |
string |
base element for the output registration matrix (highres in highres2standard.mat) |
output_bids_labels |
REQUIRED |
dictionary |
A bids filename dictionary as explained in NEEDS REFERENCE |
output_json_values |
REQUIRED |
dictionary |
Key-value pairs to additionally insert into the JSON sidecar accompanying input-to-reference transformed image |
volume |
OPTIONAL |
integer |
volume to extract using fslroi. Must specify ‘fslroi’ as true |
reference_image_params
These keys are used to identify the reference image for registration.
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
type |
REQUIRED |
string |
Type of reference: std or bids |
input_bids_location |
OPTIONAL |
string |
Input bids location: rawdata or derivatives (required if type bids) |
input_bids_labels |
OPTIONAL |
dictionary |
A bids filename dictionary as explained in NEEDS REFERENCE (required if type bids) |
output_bids_labels |
OPTIONAL |
dictionary |
Supplemental bids filename dictionary as explained in NEEDS REFERENCE (required if type bids) |
output_matrix_base |
OPTIONAL |
string |
base element for the output registration matrix (highres in highres2standard.mat) (required if type bids) |
output_json_values |
OPTIONAL |
dictionary |
Supplemental key-value pairs to additionally insert into the JSON sidecar accompanying input-to-reference transformed image (required if type bids) |
FLIRT Parameters¶
flirt_params
These parameters contains most of the inputs described in the nipype python extension manual for FSL FLIRT. See a table of our specific inputs HERE.
Optional Parameters¶
secondary_image_params
These keys should be defined if the user would like to apply the registered output to a secondary image.
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
input_bids_labels |
REQUIRED |
dictionary |
A bids filename dictionary as explained in NEEDS REFERENCE |
output_matrix_base |
REQUIRED |
string |
base element for the output registration matrix |
output_bids_labels |
REQUIRED |
dictionary |
Supplemental bids filename dictionary as explained in NEEDS REFERENCE |
standard_reference_params
These keys should be defined if the user would like to register input to a standard reference image (either )
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
file |
REQUIRED |
string |
Standard reference filename located within the FSL standard data directory |
type |
REQUIRED |
string |
type of input file: FSL CURRENTLY UNUSED |
output_matrix_suffix |
REQUIRED |
string |
suffix for the output registration matrix (standard in highres2standard.mat) |
output_bids_labels |
REQUIRED |
dictionary |
Supplemental bids filename dictionary as explained in NEEDS REFERENCE |
output_json_values |
REQUIRED |
dictionary |
Supplemental key-value pairs to additionally insert into the JSON sidecar accompanying input-to-standard transformed image |
Scan Identification JSON Control File¶
Scan Identification parameters can be found in “<project_identifier>_scan_id.json”. This file contains the inputs described in scan_id_json_table and sequence_dict_items
Sample scan_id JSON file
**Sample scan_id JSON file**
Key Name |
Required? |
Data Type |
Description |
|---|---|---|---|
|
OPTIONAL |
string |
free text to provide a brief description |
<sequence1_unique_name> |
REQUIRED |
dictionary |
scan identification parameters for sequence 1 |
<sequence2_unique_name> |
REQUIRED |
dictionary |
scan identification parameters for sequence 2 |
<sequenceN_unique_name> |
REQUIRED |
dictionary |
scan identification parameters for sequence N |
Sequence Identification Parameters
Key Name |
Required Level |
Data Type |
Description |
|---|---|---|---|
json_header |
REQUIRED |
dictionary |
Search criteria to identify NIfTI images unique to this sequence. See |
bids_labels |
REQUIRED |
dictionary |
A bids filename dictionary as explained in NEEDS REFERENCE, EXCLUDING extension key |
BidsDir |
Required |
string |
BIDS rawdata sub-directory to move the NIfTI image and associated files |
json_header search keys
Common MRI metadata fields can be found in the BIDS online specification. The specified metadata fields can be used as keys in the json_header dictionary to identify this sequence from the other acquired images as inclusionary criteria. These key-value pairs will be compared to the JSON sidecar accompanying the source NIfTI image that is created following dicom-to-nifti conversion completed either in connect_pacs_dicom_grabber or connect_dcm2nii. The values can either be an integer, string, or list[string].
Note
Exclusion criteria can be defined by preceding the keys below with “Not” (e.g., “NotProtocolName”).