CoNNECT Broad Analysis Tools¶
There are many custom analysis tools that have been developed to provide broad processing capabilities across projects, MRI scanners, imaging parameters, and processing specifications. These tools are described in this Section. Many of these tools utilize a common JSON architecture to describe project-specific inputs that are utilized by these analysis tools to allow maximum flexibility of their implementation. Specifics to these JSON control files can be found at project-specific_JSON_control_files.
Note
At this time, these functions only support command-line usage.
connect_create_project_db.py¶
This function creates the Project’s searchTable and searchSourceTable, as defined via the credentials JSON file loaded by read_credentials.py.
This function can be executed via command-line only using the following options:
$ connect_create_project_db.py -p <project_identifier>
- -p PROJECT, --project PROJECT
REQUIRED create MySQL tables named from the searchTable and searchSourceTable keys associated with the defined project
- -h, --help
show the help message and exit
- --progress
verbose mode
- -v, --version
display the current version
connect_create_raw_nii.py¶
This function moves sourcedata NIfTI files (and JSON/txt sidecars) created from connect_dcm2nii.py or convert_dicoms.py to the corresponding rawdata directory.
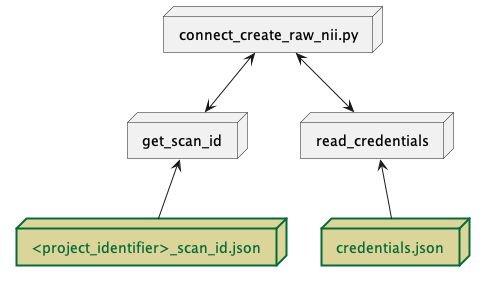
Fig. 10 Required pre-requisite files for execution of connect_create_raw_nii.py.¶
This function can be executed via command-line only using the following options:
$ connect_create_raw_nii.py -p <project_identifier> --progress --overwrite
$ connect_create_raw_nii.py -p <project_identifier> --in-dir <path to single session's sourcedata> --progress --overwrite
- -p PROJECT, --project PROJECT
REQUIRED project identifier to execute
- -i IN_DIR, --in-dir INDIR
Only execute for a single subject/session by providing a path to individual subject/session directory
- -h, --help
show the help message and exit
- --progress
verbose mode
- --overwrite
force create of rawdata files by skipping file checking
- -v, --version
display the current version
connect_dcm2nii.py¶
This function converts DICOM images to NIfTI utilizing dcm2niix and convert_dicoms.py. Files within the identified Project’s searchSourceTable are queried via MySQL for DICOM images. These DICOM images are contained within the Project’s sourcedata directory. Directories within sourcedata that contain DICOM images are then passed to dcm2niix for conversion. The NIfTI images created are then stored in the same sourcedata directory as their source DICOM directory.
See also
The dcm2niix is the most common tool for DICOM-to-NIfTI conversion, and is implemented on our Ubuntu 20.04 CoNNECT NPC nodes.
This function can be executed via command-line only:
$ connect_dcm2nii.py -p <project_identifier> --overwrite --submit --progress
- -p PROJECT, --project PROJECT
REQUIRED project to identify the associated searchSourceTable to query DICOM images for NIfTI conversion
- -h, --help
show the help message and exit
- --overwrite
force conversion by skipping directory and database checking
- --progress
verbose mode
- -s, --submit
submit conversion to the HTCondor queue for multi-threaded CPU processing
- -v, --version
display the current version
connect_flirt.py¶
This function executes registration via FSL’s FLIRT using flirt.py.
This function can be executed via command-line only:
$ connect_flirt.py -p <project_identifier> --apt --asl --struc --overwrite --progress -s
- -p PROJECT, --project PROJECT
REQUIRED project to identify the associated searchTable to query images with filenames containing BIDS labels specified in main_params.input_bids_labels
- --apt
utilize a 3D ATPw image as input for registration. This loads a FLIRT JSON control file <project_identifier>_apt_flirt_input.json
- --asl
utilize an ASL image as input for registration. This loads a FLIRT JSON control file <project_identifier>_asl_flirt_input.json
- --struc
utilize a structural image as input for registration. This loads a FLIRT JSON control file <project_identifier>_struc_flirt_input.json
- -h, --help
show the help message and exit
- --overwrite
force registration by skipping directory and database checking
- --progress
verbose mode
- -s, --submit
submit conversion to the HTCondor queue for multi-threaded CPU processing
- -v, --version
display the current version
Note
If multiple modality flags are provided (–apt, –struc, –asl), structural registration is performed first.
connect_neuro_db_query.py¶
This function will query the Project’s MySQL database using the provided search criteria defined below:
$ connect_neuro_db_query.py -p <project_identifier> -r REGEXSTR --col RETURNCOL --where SEARCHCOL --orderby ORDERBY --progress --source --opt-inclusion INCLUSION1 INCLUSION2 --opt-exclusion EXCLUSION1 EXCLUSION2 --opt-or-inclusion ORINCLUSION1 ORINCLUSION2 --version
- -p PROJECT, --project PROJECT
REQUIRED search the selected table for the indicated <project_identifier>
- -r REGEXSTR, --regex REGEXSTR
REQUIRED Search string (no wildcards, matches if the search string appears anywhere in the field specified by -w|–where)
- -h, --help
show the help message and exit
- -c RETURNCOL, --col RETURNCOL
column to return (default ‘fullpath’)
- -w SEARCHCOL, --were SEARCHCOL
column to search (default ‘filename’)
- -o ORDERBY, --orderby ORDERBY
column to sort results (default ‘fullpath’)
- --progress
verbose mode
- --source
search searchSourceTable instead of searchTable, as defined via the credentials.json file
- --opt-inclusion INCLUSION
optional additional matching search string(s) to filter results. Multiple inputs accepted through space delimiter
- --opt-exclusion EXCLUSION
optional additional exclusionary search string(s) to filter results. Multiple inputs accepted through space delimiter
- --opt-or-inclusion INCLUSION
optional additional OR matching search string(s) to filter results. Multiple inputs accepted through space delimiter
- -v, --version
display the current version
connect_neuro_db_update.py¶
This function searches the project directories to update the main (searchTable) and/or source (searchSourceTable) MySQL database tables associated with the specified project.
This function only supports command-line interface:
$ connect_neuro_db_update.py -p <project_identifier> --main --source --progress
- -p PROJECT, --project PROJECT
REQUIRED search the selected table for the indicated <project_identifier> can provide term ‘all’ to update all tables in credentials.json
- -h, --help
show the help message and exit
- --progress
verbose mode
- -s, --source
update the searchSourceTable, as defined via the credentials.json file
- -m, --main
update the searchTable, as defined via the credentials.json file
- -v, --version
display the current version
Note
This function executes nightly. This function should be executed after new files are created.
connect_pacs_dicom_grabber.py¶
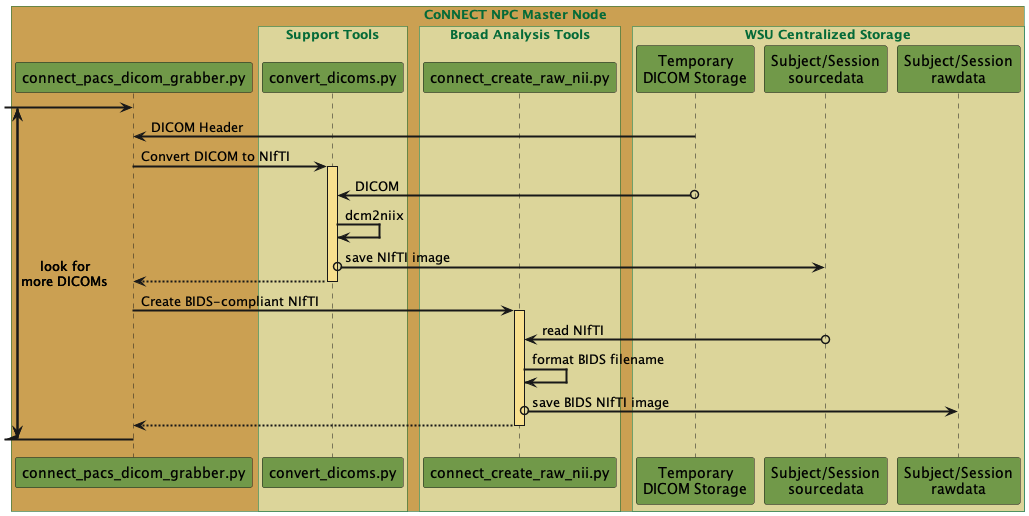
Fig. 11 Sequence diagram for the CoNNECT PACS DICOM grabber python function.¶
This function moves DICOM images from their local temporary PACS storage location (/resshare/PACS) to their associated sourcedata directory (Fig. 12), performs DICOM-to-NIfTI conversion via convert_dicoms.py, and moves the NIfTI files into a BIDS structure within the Project’s rawdata directory (Fig. 11). The <project identifier>, <subject identifier>, and <session identifer> (optional) are extracted from the DICOM header ‘Patient Name’ tag. These elements are space-delimited. The <session identifier> is appended to an acquisition date identifier using the format YYYYMMDD.

Fig. 12 Final location of source DICOM data following transfer from MRI via Orthanc PACS and connect_pacs_dicom_grabber.py.¶
Note
This function is continuously running in the background on the CoNNECT NPC master node (and loaded at startup) as the pacs-grabber.service.
This function can be executed via command-line only using the following options, HOWEVER, beware to ensure the pacs-grabber.service is stopped prior to running:
$ connect_pacs_dicom_grabber.py
- -h, --help
show the help message and exit
- -v, --version
display the current version
connect_rawdata_check.py¶
This function creates a table to indicate the absence (0) or presence (1) of MRI rawdata (NIfTI). Rawdata are identified via the project’s scan ID JSON control file. An output table in CSV format is created in the Project’s ‘processing_logs’ directory titled <project_identifier>_rawdata_check.csv.
This function can be executed via command-line only:
$ connect_rawdata_check.py -p <project_identifier>
- -p PROJECT, --project PROJECT
REQUIRED This project’s searchTable will be queried for all NIfTI images to identify images matching those scan sequences present in the scan ID JSON control file.
- -h, --help
show the help message and exit
- --progress
verbose mode
- -v, --version
display the current version