Neuro-Processing Cluster (NPC)¶
The CoNNECT NPC is a collection of systems that are centrally and locally managed. The resources are all centrally located and under the control of Wright State Computing and Telecommunications (CaTS). The CoNNECT NPC operates on a network isolated from the main WSU campus network.
Jumpbox
The jumpbox is a user-specific virtual machine (VM) running either windows or linux. The jumpbox is NIST and HIPPA compliant. A user accesses their jumpbox VM via VMware Horizon when connected to WSU’s secure network or LAN, or connected to the network via virtual private network (VPN). Users can access the remainder of the CoNNECT NPC via their jumpbox.
Master Node
The master node is the main controller of the CoNNECT NPC. Users access the master node via secure shell (SSH) connections through their jumpbox. The master node has 40 physical cores and 756GB RAM, and is running Ubuntu 20.04.
Core Node(s)
Core nodes are the workhorse of the CoNNECT NPC. The core nodes run the same operating system as the master, Ubuntu 20.04. Currently, there is a single core node with 40 physical cores and 756GB RAM. Processing can be conducted utilizing the core node(s) by supplying the submit option to the programs described within this manual
Open-Source Software¶
Picture Archiving and Communication System (PACS)¶
Orthanc is the official open-source PACS system in place at the WSU CoNNECT. Using this system, which runs on the master node of the CoNNECT NPC, DICOM images can be sent directly from the MRI scanner to the CoNNECT NPC centralized storage. The PACS Grabber Service then automatically detects new dicoms arriving to the PACS storage location, moves them to the appropriate project/subject/session’s sourcedata directory, converts them to NIfTI files and stores them in the project/subject/session’s rawdata directory according to BIDS specifications.
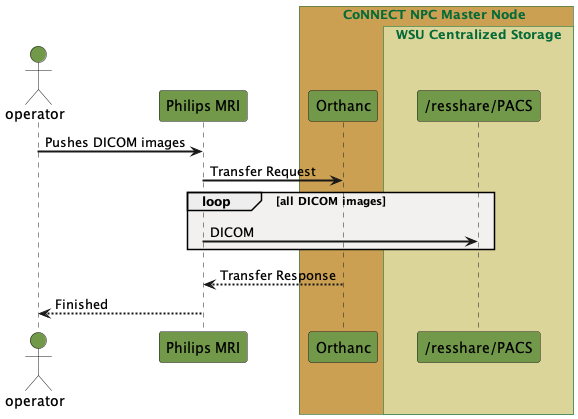
Fig. 9 PACS data transfer from MRI to centralized storage via CoNNECT NPC.¶
Image Viewing and Analysis Software¶
AFNI¶
AFNI is an NIH developed suite to visualize and analyze functional imaging.
The afni package (installed from http://neuro.debian.net/pkgs/afni.html) is located at /usr/share/afni.
AFNI is commonly utilized to overlay fMRI statistics from FSL FEAT or other output on top of T1-weighted images. Through AFNI surface mapper (SUMA), freesurfer inflated volumes can be used as the background image for the statistics, which can be viewed in 3D, as well as many other views. With certain datasets, this can provide powerful visuals.
$ afni
dcm2niix¶
Dcm2niix is a conversion software package developed at Chris Rorden’s Neuropsychology Lab at University of South Carolina and is the distributed by NeuroImaging and Resources Collaboratory (NITRC).
A copy of dcm2niix has been compiled and placed in the scratch ‘bash’ folder, /mnt/ss_rhb1/scratch/bash. This path has been added to the system path so it can be ran from any directory without the need to know the location. A copy is also in the ubuntu user download folder.
The function dcm2niix converts raw DICOM images to NIfTI (or other) formats. The function should be executed with the flags specified in Table 4.
$ dcm2niix
Freesurfer¶
Freesurfer Freesurfer is developed by Massachusetts General Hospital/Harvard University to analyze MRI images of the brain. The current version is 7.1.1; however, version 6 is installed.
The freesurfer package (installed from ) is located at /usr/share/freesurfer. This path is stored in the FREESURFER_DIR variable available in bash and c-shell terminals.
common functions:
Main freesurfer command to begin the cortical reconstruction process. The entire process takes 12-24hrs to complete. Preferred command line arguments are provided in Table 3.
Note
CREATE TABLE
$ recon-all
The volume and surface viewer to assess quality of cortical reconstruction.
$ freeview
FMRIB Software Library (FSL)¶
FSL FMRIB Software Library (FSL) is a library of fMRI, MRI and DTI analysis tools developed by Oxford University, UK. The current installed version is 6.0.4.
The fsl package is located at /usr/share/fsl/6.0. This path is stored in the FSL_DIR variable available in bash and c-shell terminals.
Common Functions
$ fsl
Main GUI for FSL. This GUI can be used to create individual and higher-level designs for fMRI analyses, execute individual or higher-level designs of fMRI analyses, run brain extraction, process DTI, perform registration, perform ICA for resting-state fMRI, or generate simulated MRI scans.
See also
Details can be found here.
$ fsleyes
MRI visualization GUI.
See also
The user guide can be found here.
$ feat
Command line tool to execute fMRI design files.
See also
More info can be found here.
$ randomise
Randomise (and randomise_parallel) is a non-parametric permutation inference tool for neuroimaging data. Randomise utilizes modelling and inferences using standard designs as used in FEAT. The main pitfall of randomize is the accommodation of correlated datasets (repeated measures). However, some cases of repeated measures can be accommodated.
$ oxford_asl
oxford_asl is a command line utility that can quantify cerebral perfusion (CBF) data from ASL, including motion-correction, registration, partial volume correction, and distortion correction.
See also
Command line user guide for oxford asl or BASIL, the GUI-based version of oxford_asl.
LCModel¶
Note
To be developed
MySQL Database¶
A CoNNECT MySQL database has been implemented on the master node to index files in each project’s directory. This database and associated tools therein are local to only the master node and, thus, core nodes do not have the ability to query or update. Different tables must be established for each independent project, which can be developed using this custom toolkit.
MySQL can be accessed through the command line:
$ mysql --login-path=client <database>
where <database> is replaced with the MySQL database. Currently, all projects are contained in a single database, “CoNNECT”.
Each project will have two tables in the CoNNECT database: one main table containing all files except source files and one source table containing files within the BIDS sourcedata directory. The main table (referred to in the project’s credentials as searchTable, see credentials.json file) contains the elements described in Table 2. The sourcedata table contains the elements described in Table 3.
MySQL allows efficent queries of files contained within a project’s directories. This will optimize file searching and data processing. Any ‘-‘ are illegal characters in the table name and are generally replaced with an underscore (‘_’).
Column Name |
Column Description |
Char Size |
|---|---|---|
fullpath |
Full path to the local file, including filename and extension |
255 |
filename |
Filename and extension, excluding the path |
255 |
basename |
Filename, excluding extension and path. NULL if no basename |
255 |
extension |
Extension, excluding filename and path. NULL if no extension |
48 |
Column Heading |
Column Description |
Char Size |
|---|---|---|
fullpath |
Full path to the local file, including filename and extension |
255 |
filename |
Filename and extension, excluding the path |
255 |
Helpful MySQL Commands¶
These helpful commands can be executed on the master node:
Logging in to MySQL¶
$ mysql --login-path=client <database>
Exiting MySQL¶
$ exit;
Creating a New Database¶
$ CREATE DATABASE <database_name>;
Creating a New Table¶
$ CREATE TABLE <table_name> (<column1_name> <column1_size> <column2_name> <column2_size>);
List All Tables in Database¶
$ SHOW tables;
Retrieve ALL Columns from a Table¶
$ SELECT * FROM <table_name>;
Retrieve ALL Columns from a Table Matching String¶
$ CREATE DATABASE <database_name> WHERE <column> REGEXP “<search_string>”;
Determine Last Update Time for a Table¶
$ SELECT UPDATE_TIME FROM information_schema.tables WHERE TABLE_SCHEMA = <database> AND TABLE_NAME = <table>;