5.3 Protein Interactions Modulated by Chemical Energy: Actin, Myosin, and Molecular Motors
Organisms move. Cells move. Organelles and macromolecules within cells move. Most of these movements arise from the activity of a fascinating class of protein-based molecular motors. Fueled by chemical energy, usually derived from ATP, large aggregates of motor proteins undergo cyclic conformational changes that accumulate into a unified, directional force — the tiny force that pulls apart chromosomes in a dividing cell, and the immense force that levers a pouncing, quarter-ton jungle cat into the air.
The interactions among motor proteins, as you might predict, feature complementary arrangements of ionic, hydrogen-bonding, and hydrophobic groups at protein binding sites. In motor proteins, however, the resulting interactions achieve exceptionally high levels of spatial and temporal organization.
Motor proteins underlie the migration of organelles along microtubules, the motion of eukaryotic and bacterial flagella, and the movement of some proteins along DNA. Here, we focus on the well-studied example of the contractile proteins of vertebrate skeletal muscle as a paradigm for how proteins translate chemical energy into motion.
The Major Proteins of Muscle Are Myosin and Actin
The contractile force of muscle is generated by the interaction of two proteins, myosin and actin. These proteins are arranged in filaments that undergo transient interactions and slide past each other to bring about contraction. Together, actin and myosin make up more than 80% of the protein mass of muscle.
Myosin ( 520,000) has six subunits: two heavy chains (each of 220,000) and four light chains (each of 20,000). The heavy chains account for much of the overall structure. At their carboxyl termini, they are arranged as extended α helices, wrapped around each other in a fibrous, left-handed coiled coil similar to that of α-keratin (Fig. 5-26). At its amino terminus, each heavy chain has a large globular domain containing a site where ATP is hydrolyzed. The light chains are associated with the globular domains.
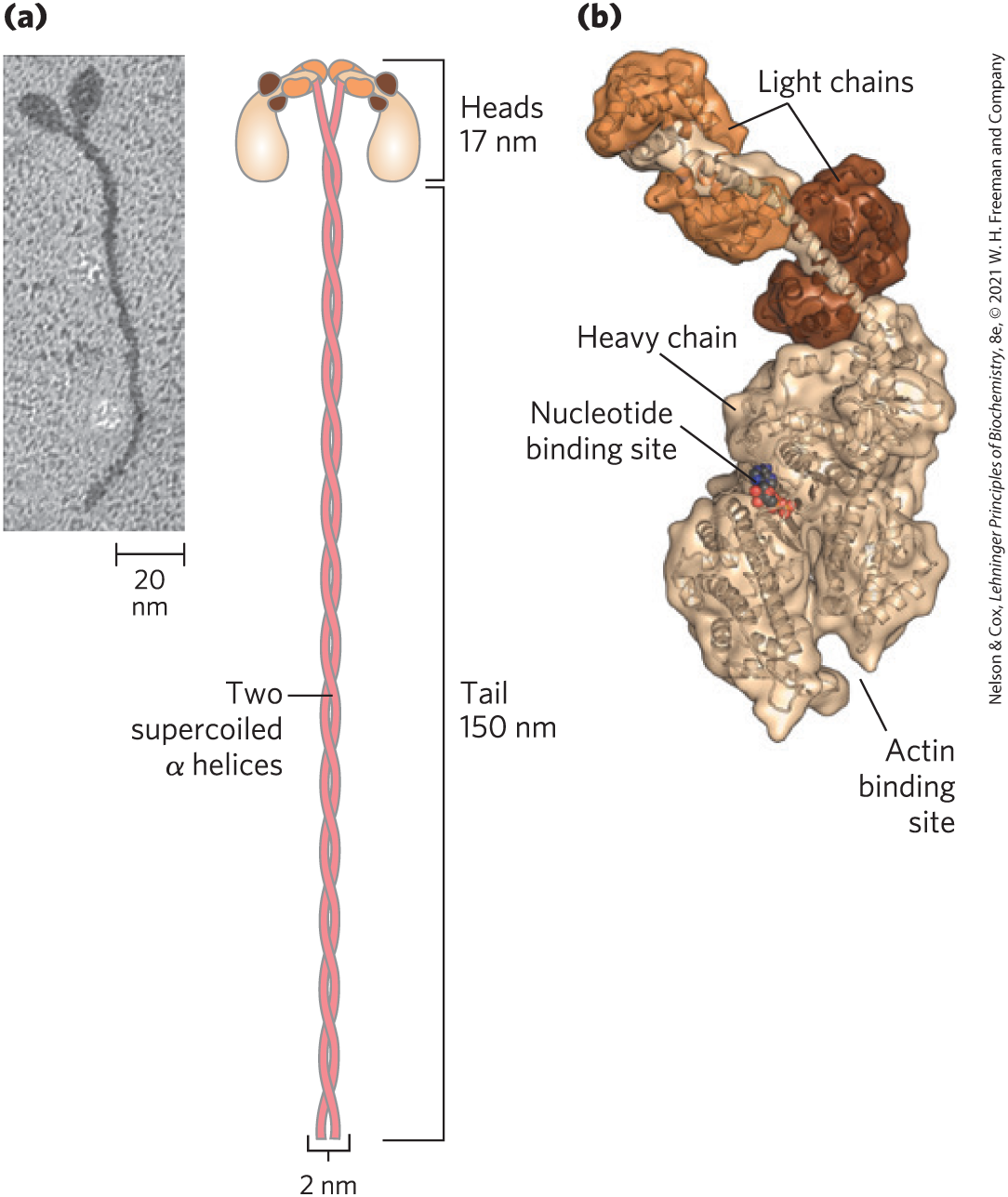
FIGURE 5-26 Myosin. (a) Myosin has two heavy chains: the carboxyl termini forming an extended coiled coil tail and the amino termini having clublike globular head domains. Two myosin light chains are associated with each myosin head. (b) A three-dimensional representation of the myosin head, showing the binding sites for nucleotide (ATP) and actin. [(a) Research from Takeshi Katayama, et al. “Stimulatory effects of arachidonic acid on myosin ATPase activity and contraction of smooth muscle via myosin motor domain,” Am. J. Physiol. Heart Circ. Physiol. Vol 298, Issue 2, pp. H505–H514, February 2010, Fig. 6b. (b) Data from PDB ID 2MYS, I. Rayment et al., Science 261:50, 1993.]
In muscle cells, molecules of myosin aggregate to form structures called thick filaments (Fig. 5-27a). These rodlike structures are the core of the contractile unit. Within a thick filament, several hundred myosin molecules are arranged with their fibrous “tails” associated to form a long, bipolar structure. The globular domains project from either end of this structure, in regular stacked arrays.
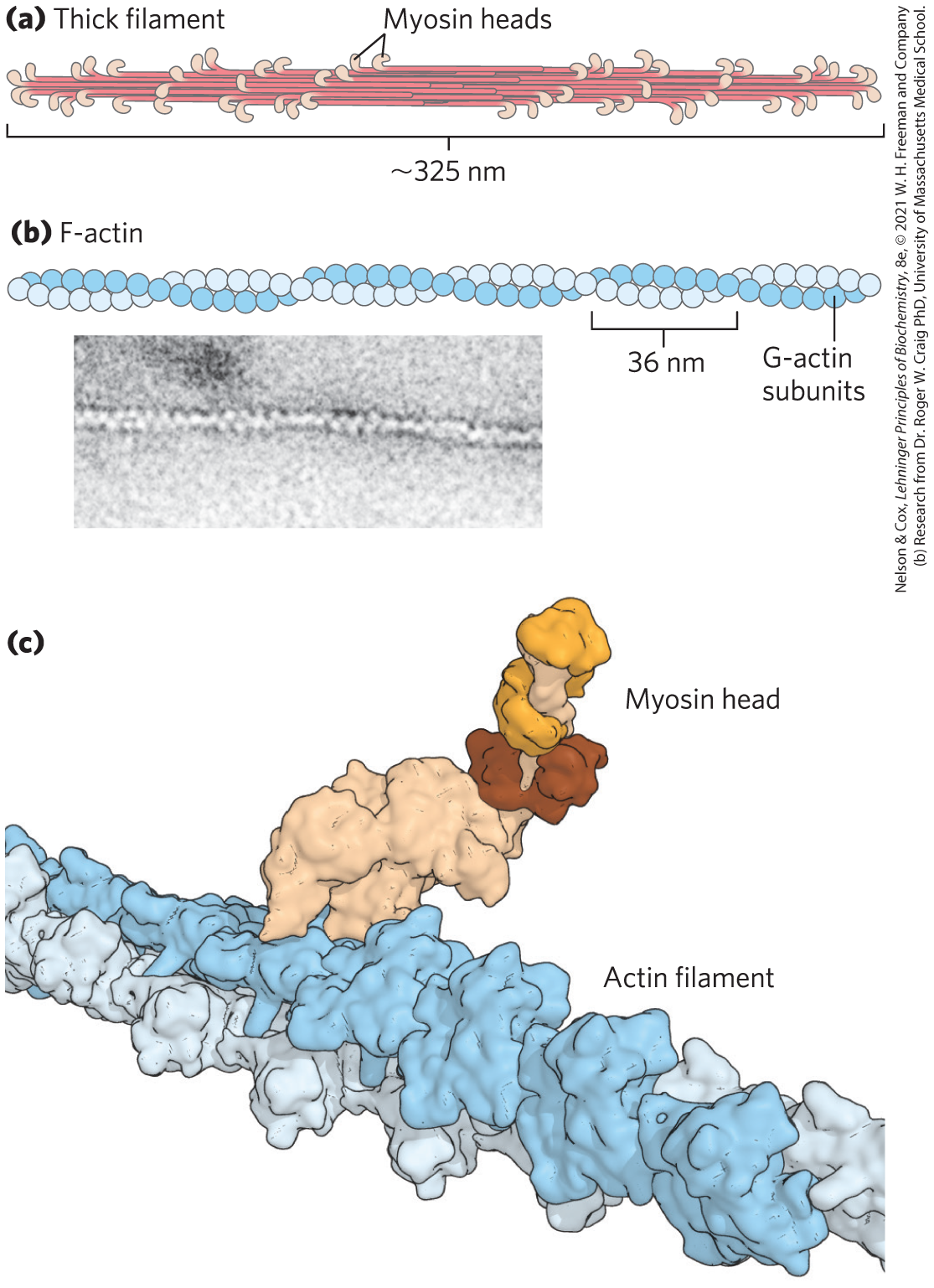
FIGURE 5-27 The major components of muscle. (a) Myosin aggregates to form a bipolar structure called a thick filament. (b) F-actin is a filamentous assemblage of G-actin monomers that polymerize two by two, giving the appearance of two filaments spiraling about one another in a right-handed fashion. (c) Space-filling model of an actin filament with one myosin head bound to an actin monomer within the filament. [(c) Data from PDB ID 2MYS, I. Rayment et al., Science 261:50, 1993; PDB ID 6BNQ, P. S. Gurel et al., eLife 6, 2017, doi 10.7554/eLife.31125]
The second major muscle protein, actin, is abundant in almost all eukaryotic cells. In muscle, molecules of monomeric actin, called G-actin (globular actin; 42,000), associate to form a long polymer called F-actin (filamentous actin). The thin filament consists of F-actin (Fig. 5-27b), along with the proteins troponin and tropomyosin (discussed below). The filamentous parts of thin filaments assemble as successive monomeric actin molecules add to one end. Upon addition, each monomer binds ATP, then hydrolyzes it to ADP, so every actin molecule in the filament is complexed to ADP. This ATP hydrolysis by actin functions only in the assembly of the filaments; it does not contribute directly to the energy expended in muscle contraction. Each actin monomer in the thin filament can bind tightly and specifically to one myosin head group (Fig. 5-27c).
Additional Proteins Organize the Thin and Thick Filaments into Ordered Structures
Skeletal muscle consists of parallel bundles of muscle fibers, each fiber a single, very large, multinucleated cell, 20 to 100 μm in diameter, formed from many cells fused together; a single fiber often spans the length of the muscle. Each fiber contains about 1,000 myofibrils, 2 μm in diameter, each consisting of a vast number of regularly arrayed thick and thin filaments complexed to other proteins (Fig. 5-28). A system of flat membranous vesicles called the sarcoplasmic reticulum surrounds each myofibril. Examined under the electron microscope, muscle fibers reveal alternating regions of high electron density and low electron density, called the A bands and I bands (Fig. 5-28b, c). The A and I bands arise from the arrangement of thick and thin filaments, which are aligned and partially overlapping. The I band is the region of the bundle that in cross section would contain only thin filaments. The darker A band stretches the length of the thick filament and includes the region where parallel thick and thin filaments overlap. Bisecting the I band is a thin structure called the Z disk, perpendicular to the thin filaments and serving as an anchor to which the thin filaments are attached. The A band, too, is bisected by a thin line, the M line or M disk, a region of high electron density in the middle of the thick filaments. The entire contractile unit, consisting of bundles of thick filaments interleaved at either end with bundles of thin filaments, is called the sarcomere. The arrangement of interleaved bundles allows the thick and thin filaments to slide past each other (by a mechanism discussed below), causing a progressive shortening of each sarcomere (Fig. 5-28d).
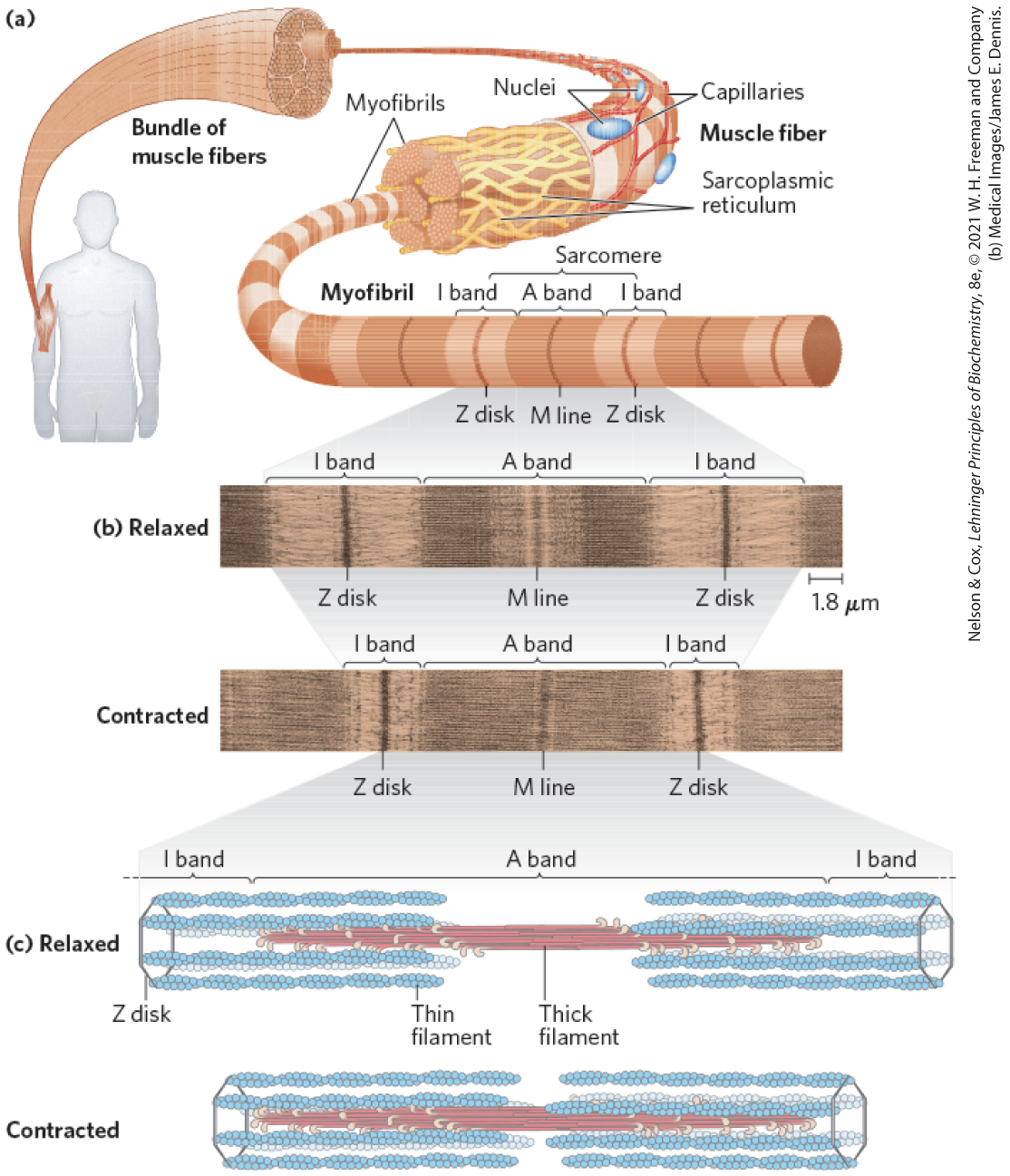
FIGURE 5-28 Skeletal muscle. (a) Muscle fibers consist of single, elongated, multinucleated cells that arise from the fusion of many precursor cells. The fibers are made up of many myofibrils (only six are shown here for simplicity) surrounded by the membranous sarcoplasmic reticulum. The organization of thick and thin filaments in a myofibril gives it a striated appearance. When muscle contracts, the I bands narrow and the Z disks move closer together, as seen in electron micrographs of (b) relaxed muscle and (c) contracted muscle. (d) Thick filaments are bipolar structures created by the association of many myosin molecules. Muscle contraction occurs by the sliding of the thick and thin filaments past each other so that the Z disks in neighboring I bands draw closer together. The thick and thin filaments are interleaved such that each thick filament is surrounded by six thin filaments.
Myosin Thick Filaments Slide along Actin Thin Filaments
The interaction between actin and myosin, like that between all proteins and ligands, involves weak bonds. When ATP is not bound to myosin, a face on the myosin head group binds tightly to actin (Fig. 5-29). When ATP binds to myosin and is hydrolyzed to ADP and phosphate, a coordinated and cyclic series of conformational changes occurs in which myosin releases the F-actin subunit and binds another subunit farther along the thin filament.
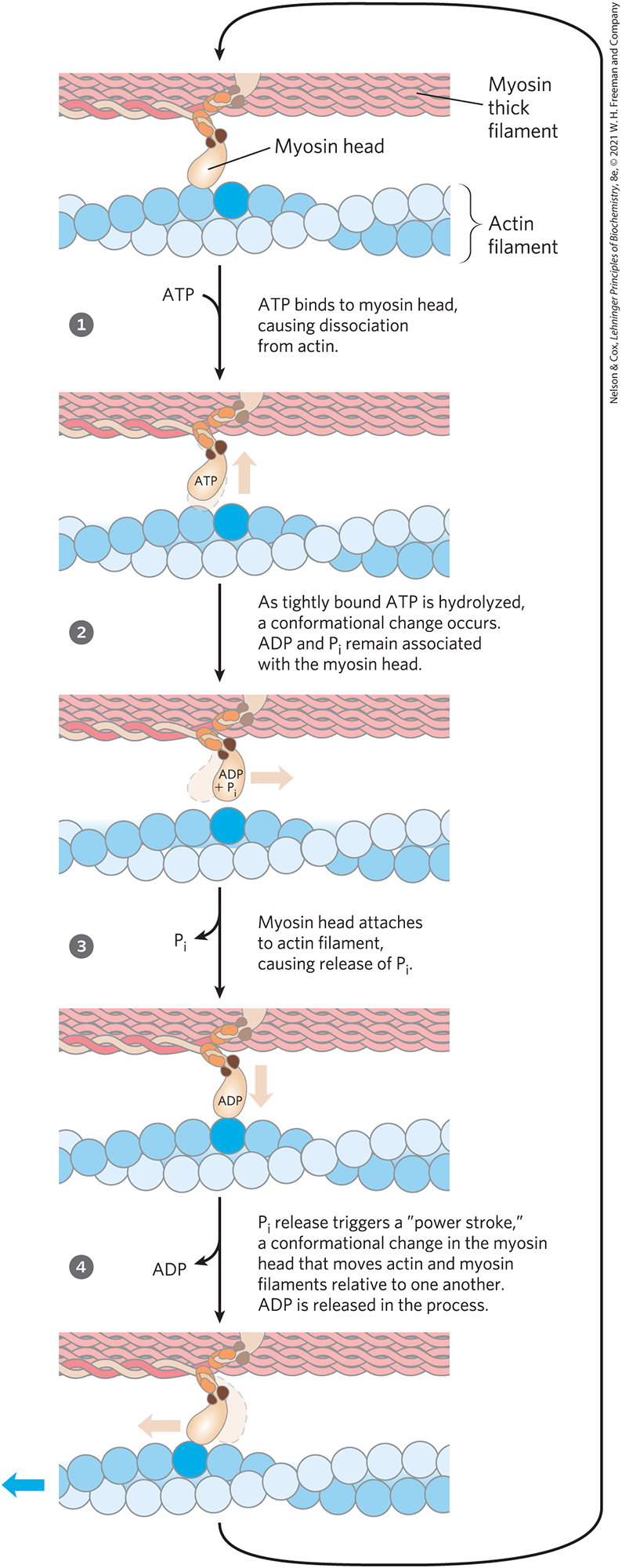
FIGURE 5-29 Molecular mechanism of muscle contraction. Conformational changes in the myosin head that are coupled to stages in the ATP hydrolytic cycle cause myosin to successively dissociate from one actin subunit, then associate with another farther along the actin filament. In this way, the myosin heads slide along the thin filaments, drawing the thick filament array into the thin filament array (see Fig. 5-28).
The cycle has four major steps (Fig. 5-29). In step , ATP binds to myosin and a cleft in the myosin molecule opens, disrupting the actin-myosin interaction so that the bound actin is released. ATP is then hydrolyzed in step , causing a conformational change in the protein to a “high-energy” state that moves the myosin head and changes its orientation in relation to the actin thin filament. Myosin then binds weakly to an F-actin subunit closer to the Z disk than the one just released. As the phosphate product of ATP hydrolysis is released from myosin in step , another conformational change occurs in which the myosin cleft closes, strengthening the myosin-actin binding. This is followed quickly by step , a “power stroke” during which the conformation of the myosin head returns to the original resting state, its orientation relative to the bound actin changing so as to pull the tail of the myosin toward the Z disk. ADP is then released to complete the cycle. Each cycle generates about 3 to 4 pN (piconewtons) of force and moves the thick filament 5 to 10 nm relative to the thin filament.
Because there are many myosin heads in a thick filament, at any given moment some (probably 1% to 3%) are bound to thin filaments. This prevents thick filaments from slipping backward when an individual myosin head releases the actin subunit to which it was bound. The thick filament thus actively slides forward past the adjacent thin filaments. This process, coordinated among the many sarcomeres in a muscle fiber, brings about muscle contraction.
The interaction between actin and myosin must be regulated so that contraction occurs only in response to appropriate signals from the nervous system. The regulation is mediated by a complex of two proteins, tropomyosin and troponin (Fig. 5-30). Tropomyosin binds to the thin filament, blocking the attachment sites for the myosin head groups. Troponin is a -binding protein. A nerve impulse causes release of ions from the sarcoplasmic reticulum. The released binds to troponin (another protein-ligand interaction) and causes a conformational change in the tropomyosin-troponin complexes, exposing the myosin-binding sites on the thin filaments. Contraction follows.
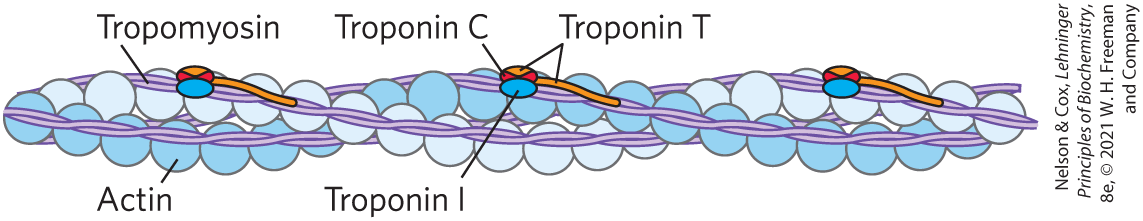
FIGURE 5-30 Regulation of muscle contraction by tropomyosin and troponin. Tropomyosin and troponin are bound to F-actin in the thin filaments. In the relaxed muscle, these two proteins are arranged around the actin filaments so as to block the binding sites for myosin. Tropomyosin is a two-stranded coiled coil of α helices, the same structural motif as in α-keratin (see Fig. 4-11). It forms head-to-tail polymers twisting around the two actin chains. Troponin is attached to the actin-tropomyosin complex at regular intervals of 38.5 nm. Troponin consists of three different subunits: I, C, and T. Troponin I prevents binding of the myosin head to actin; troponin C has a binding site for ; and troponin T links the entire troponin complex to tropomyosin. When the muscle receives a neural signal to initiate contraction, is released from the sarcoplasmic reticulum (see Fig. 5-28a) and binds to troponin C. This causes a conformational change in troponin C, which alters the positions of troponin I and tropomyosin so as to relieve the inhibition by troponin I and allow muscle contraction.
Working skeletal muscle requires two types of molecular functions that are common in proteins: binding and catalysis. The actin-myosin interaction, a protein-ligand interaction like that of immunoglobulins with antigens, is reversible and leaves the participants unchanged. This interaction illustrates why reversibility is important; a permanent actin-myosin interaction defines the state of rigor mortis, a state we all want to avoid as long as possible. When ATP binds myosin, however, it is hydrolyzed to ADP and (inorganic phosphate). Myosin is not only an actin-binding protein, but also an ATPase — an enzyme. The function of enzymes in catalyzing chemical transformations is the topic of the next chapter.
SUMMARY 5.3 Protein Interactions Modulated by Chemical Energy: Actin, Myosin, and Molecular Motors
- Protein-ligand interactions achieve a special degree of spatial and temporal organization in motor proteins. Muscle contraction results from choreographed interactions between myosin and actin, coupled to the hydrolysis of ATP by myosin.
- Myosin consists of two heavy chains and four light chains, forming a fibrous coiled coil (tail) domain and a globular (head) domain. Myosin molecules are organized into thick filaments, which slide past thin filaments composed largely of actin. ATP hydrolysis in myosin is coupled to a series of conformational changes in the myosin head, leading to dissociation of myosin from one F-actin subunit and its eventual reassociation with another, farther along the thin filament. The myosin thus slides along the actin filaments.
- Muscle contraction is stimulated by the release of from the sarcoplasmic reticulum. The binds to the protein troponin, leading to a conformational change in a troponin-tropomyosin complex that triggers the cycle of actin-myosin interactions.
 The interaction between actin and myosin, like that between all proteins and ligands, involves weak bonds. When ATP is not bound to myosin, a face on the myosin head group binds tightly to actin (
The interaction between actin and myosin, like that between all proteins and ligands, involves weak bonds. When ATP is not bound to myosin, a face on the myosin head group binds tightly to actin ( , ATP binds to myosin and a cleft in the myosin molecule opens, disrupting the actin-myosin interaction so that the bound actin is released. ATP is then hydrolyzed in step
, ATP binds to myosin and a cleft in the myosin molecule opens, disrupting the actin-myosin interaction so that the bound actin is released. ATP is then hydrolyzed in step  , causing a conformational change in the protein to a “high-energy” state that moves the myosin head and changes its orientation in relation to the actin thin filament. Myosin then binds weakly to an F-actin subunit closer to the Z disk than the one just released. As the phosphate product of ATP hydrolysis is released from myosin in step
, causing a conformational change in the protein to a “high-energy” state that moves the myosin head and changes its orientation in relation to the actin thin filament. Myosin then binds weakly to an F-actin subunit closer to the Z disk than the one just released. As the phosphate product of ATP hydrolysis is released from myosin in step  , another conformational change occurs in which the myosin cleft closes, strengthening the myosin-actin binding. This is followed quickly by step
, another conformational change occurs in which the myosin cleft closes, strengthening the myosin-actin binding. This is followed quickly by step  , a “power stroke” during which the conformation of the myosin head returns to the original resting state, its orientation relative to the bound actin changing so as to pull the tail of the myosin toward the Z disk. ADP is then released to complete the cycle. Each cycle generates about 3 to 4 pN (piconewtons) of force and moves the thick filament 5 to 10 nm relative to the thin filament.
, a “power stroke” during which the conformation of the myosin head returns to the original resting state, its orientation relative to the bound actin changing so as to pull the tail of the myosin toward the Z disk. ADP is then released to complete the cycle. Each cycle generates about 3 to 4 pN (piconewtons) of force and moves the thick filament 5 to 10 nm relative to the thin filament. Protein-ligand interactions achieve a special degree of spatial and temporal organization in motor proteins. Muscle contraction results from choreographed interactions between myosin and actin, coupled to the hydrolysis of ATP by myosin.
Protein-ligand interactions achieve a special degree of spatial and temporal organization in motor proteins. Muscle contraction results from choreographed interactions between myosin and actin, coupled to the hydrolysis of ATP by myosin.