fMRIPrep
| Dependency | Purpose |
|---|---|
| FSL (FLIRT, FNIRT, MCFLIRT, SUSAN, FAST) | Motion correction, skull stripping, bias field correction, nonlinear warping |
| ANTs (Advanced Normalization Tools) | Spatial normalization (T1→MNI), bias field correction |
| FreeSurfer | Cortical surface reconstruction (optional, but recommended) |
| AFNI (3dTshift, 3dSkullStrip) | Slice-timing correction, alternative skull stripping |
| Workflows from Nilearn & Nipype | Pipeline orchestration and image processing |
| NumPy, SciPy, NiBabel, pandas | Data processing |
| Matplotlib, seaborn | Quality control (QC) reporting |
| TemplateFlow | Standardized brain templates |
FSL::feat
Outputs :
First-Level Analysis ( Single-Subject )
GLM Parameter Estimates (
cope1.nii.gz,varcope1.nii.gz)T-Statistics & Z-Statistics (
tstat1.nii.gz,zstat1.nii.gz)Design Matrix (
design.mat,design.con,design.fsf)Motion Outliers Report (
motion_outliers.txt)Contrast Maps ( e.g.,
zstat1.nii.gzfor statistical significance )Registration Outputs ( e.g.,
example_func2highres.matfor aligning functional to anatomical )
Higher-Level Analysis ( Group-Level )
Group statistics ( mixed-effects modeling )
Cluster-corrected results
Thresholded statistical maps
fMRIPrep Outputs Needed for Nilearn
| fMRIPrep Output | Used for |
|---|---|
sub-*_task-*_bold_space-MNI152NLin2009cAsym_preproc.nii.gz | Preprocessed functional images |
sub-*_desc-brain_mask.nii.gz | Brain mask for extracting signal |
sub-*_desc-confounds_timeseries.tsv | Motion parameters, CompCor regressors |
sub-*_space-MNI152NLin2009cAsym_res-2_desc-preproc_bold.nii.gz | Smoothed images (if needed) |
sub-*_from-T1w_to-MNI152_desc-linear_xfm.h5 | Transformation matrices (for custom ROI masking) |
Nilearn Equivalent of FEAT Steps
| FEAT Step | Nilearn Alternative |
|---|---|
| Motion Correction | Already handled by fMRIPrep |
| Spatial Normalization | Already handled by fMRIPrep |
| Confound Regression | nilearn.image.clean_img() |
| GLM First-Level | nilearn.glm.first_level.FirstLevelModel() |
| Statistical Maps (t-stats, z-stats) | .compute_contrast() in FirstLevelModel |
| ROI Extraction | nilearn.input_data.NiftiMasker |
| MVPA/Decoding | nilearn.decoding.SearchLight |
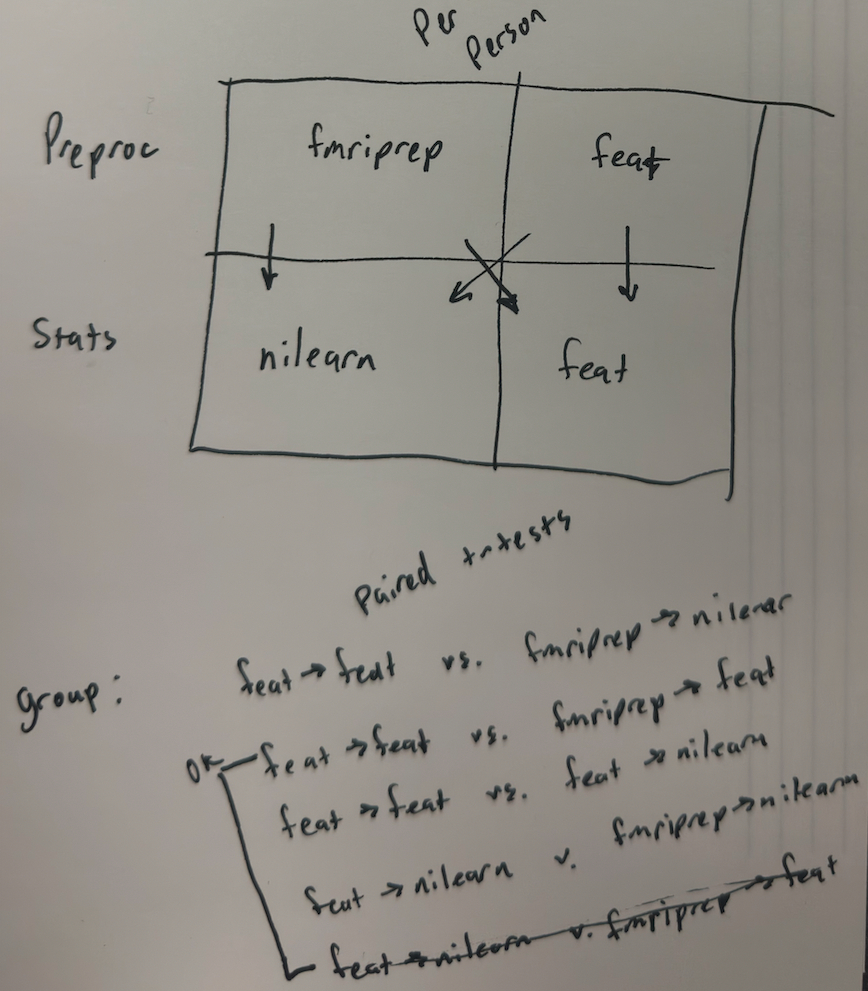
Per Person
Option 1 : fMRIPrep ➡️ FSL::feat
Run fMRIPrep: Prepares cleaned functional images with confound regressors.
Import into FEAT: Use
*_preproc_bold.nii.gzas input for FEAT.Apply Confound Regressors: Use
desc-confounds_timeseries.tsvin FEAT model.Run First-Level Analysis: Apply GLM to analyze task-based activity.
Higher-Level Analysis: Combine across subjects.
Option 2 : fMRIPrep ➡️ nilearn
Run fMRIPrep: Prepares standardized, preprocessed fMRI data.
Load in Nilearn: Use
nibabelandnilearn.imageto load preprocessed outputs.Apply Masking & Confound Removal: Use
nilearn.input_data.NiftiMaskerand regress out motion, CompCor signals, etc.Perform Statistical Modeling: Use
nilearn.glm.first_level.FirstLevelModelinstead of FSL FEAT.Extract ROIs & Perform Decoding: Use
nilearn.decodingfor MVPA, ornilearn.connectomefor network analyses.
Option 3 : FSL::feat ➡️ nilearn
Run FEAT (First-Level Analysis): Process individual subject-level fMRI data.
Extract Statistical Maps: Use
stats/cope1.nii.gz,stats/zstat1.nii.gz, etc.Load in Nilearn: Use
nibabelandnilearn.image.load_img()to read statistical maps.Perform Further Analysis: Apply machine learning, MVPA, or advanced modeling using
nilearn.decodingornilearn.glm.Visualization & Interpretation: Use
nilearn.plottingfor displaying activation maps.
Option 4 : FSL::feat ➡️ FSL:feat
Run FEAT (First-Level Analysis): Process individual subject fMRI data.
Extract First-Level Outputs: Obtain contrast maps (
cope.nii.gz), variance maps (varcope.nii.gz), and statistical results.Register to Standard Space: Use
example_func2standard.matto align functional data.Set Up Group-Level Model: Create a
design.matanddesign.confile for group-level GLM analysis.Run Higher-Level FEAT Analysis: Apply mixed-effects modeling to combine subject data.
Correct for Multiple Comparisons: Use cluster-based thresholding or TFCE correction.
Generate Final Statistical Maps: Export
thresh_zstat.nii.gzfor interpretation.
Group Level
FSL::feat Group-Level
Collect First-Level Outputs: Gather
cope.nii.gzandvarcope.nii.gzfiles from each subject.Prepare a Group Design Matrix: Create a
design.matanddesign.confor higher-level analysis.Run Higher-Level FEAT: Perform mixed-effects analysis to model group differences.
Apply Statistical Thresholding: Use FSL's thresholding options (cluster-based correction, TFCE, etc.).
Interpret Results: Review
thresh_zstat.nii.gzfor significant group-level activations.
Nilearn Group-Level
Load Individual Subject Maps: Use
nilearn.image.load_img()to import first-level statistical maps.Apply Standardization: Use
nilearn.image.mean_img()ornilearn.image.math_img()for normalization.Perform Group-Level GLM: Use
nilearn.glm.second_level.SecondLevelModel()for a random-effects model.Define Contrasts: Specify conditions or groups for comparison.
Run Statistical Analysis: Compute group contrasts and extract statistical maps.
Multiple Comparison Correction: Use
nilearn.stats.threshold_stats_img()for FDR correction.Visualize Results: Use
nilearn.plotting.plot_stat_map()to render group-level activation.