Cell Communication - Receptors
1. What are the four main receptor subtypes organized fastest to slowest?
Ligand-Gated Ion Channels ( ionotropic ) : Fastest response time ( milliseconds ); involved in rapid synaptic signaling.
G-Protein Coupled Receptors ( GPCRs ) ( metabotropic ) : Slower than ion channels ( seconds ); activate intracellular signaling pathways.
Enzyme-Coupled Receptors: Slower response ( minutes ); often involve kinase activity leading to phosphorylation cascades.
Nuclear / Intracellular Receptors: Slowest response ( minutes to hours ); typically involve changes in gene expression by interacting with DNA.
2. What do we mean by affinity and efficacy?
Affinity: The ability of a drug to bind to its receptor. High-affinity drugs bind effectively at lower concentrations.
Efficacy: The ability of a drug-receptor complex to produce a physiological response. A drug can have high affinity but low efficacy.
3. How can we measure affinity?
Radioligand Binding Assays: Utilize radiolabeled drugs to measure how well a drug binds to receptors in tissue samples.
Prepare the Tissue/Cell Membrane Sample:
Homogenize the tissue or prepare membrane fractions from cultured cells that express the receptor of interest. This step ensures the receptors are available for binding with the radiolabeled ligand.
Incubate with Radiolabeled Ligand:
Incubate the tissue or membrane preparation with a radiolabeled version of the drug (ligand) that binds to the receptor. This step allows the drug to interact with the receptor.
Measure Total Binding:
After incubation, filter the mixture to separate bound ligand from free ligand. Wash the filters to remove unbound radioligand and measure the radioactivity in the bound fraction (this is total binding, which includes both specific and non-specific binding).
Measure Non-Specific Binding:
Perform a separate assay where you add an excess of a known competitor drug (usually an antagonist or non-radioactive ligand) that binds to the receptor but is not radiolabeled. This will occupy all the specific binding sites on the receptor, so any remaining binding measured is non-specific (e.g., the radioligand sticking to other parts of the tissue, non-receptor sites, etc.).
Increase Concentration of Radioligand:
Conduct the assay across a range of radioligand concentrations to observe how binding changes. This helps in creating a binding curve to quantify affinity.
Subtract Non-Specific Binding to Get Specific Binding:
Subtract the non-specific binding measured in step 4 from the total binding measured in step 3. The difference gives the specific binding, which represents the radioligand binding specifically to the receptor of interest.
Analyze Data:
Use the data to plot a saturation binding curve or perform a Scatchard plot analysis. This allows you to calculate key parameters like the dissociation constant
Non-specific binding: This is measured by adding a high concentration of a non-radioactive competitor drug.
Specific binding: This is calculated by subtracting non-specific binding from total binding.
Affinity: The strength of binding is typically expressed by the dissociation constant
The dissociation constant
Left shift on velocity vs concentration curve = more affinity
Right shift on velocity vs concentration curve = less affinity
Let some binding event / reaction of Agonist ( A ) and Receptor ( R ) :
4. What is an agonist and what is an antagonist?
Agonists: binds to a receptor = produces a response
Example = fentanyl
Antagonists: binds to the same receptor = doesn't produce a response
Example = narcan
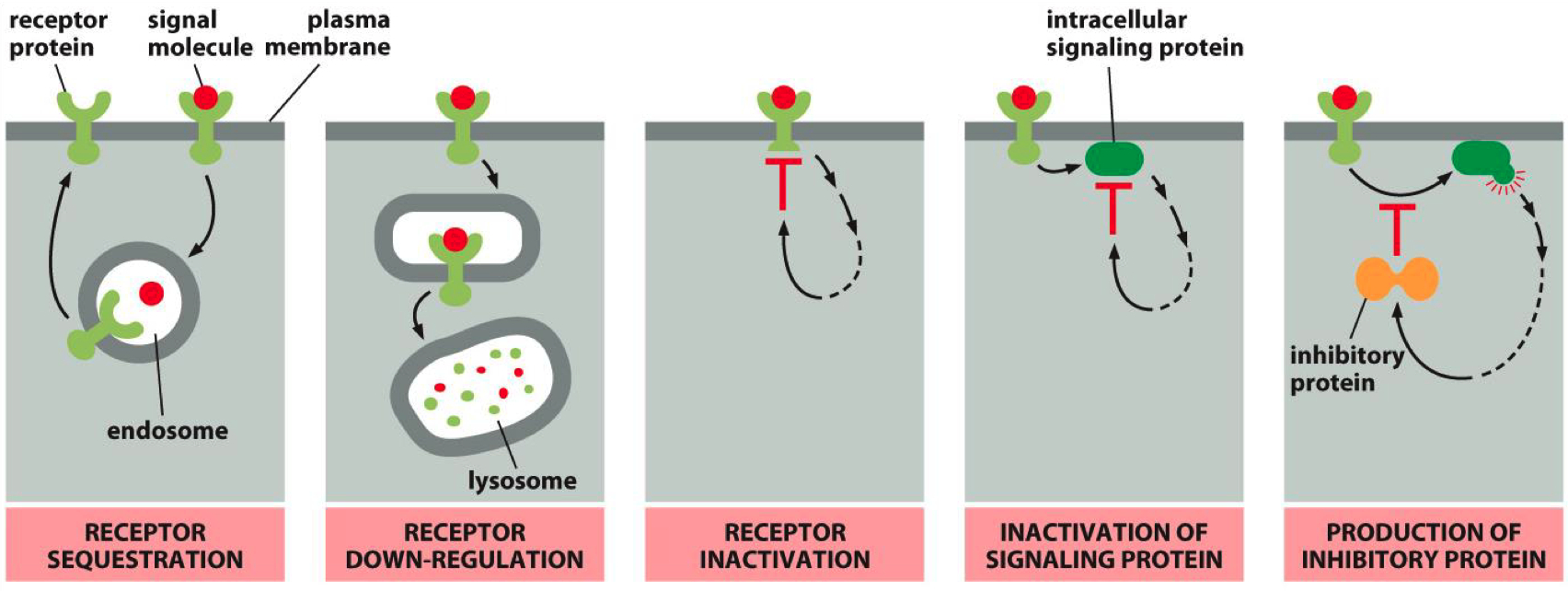
5. Describe mechanisms that can inactivate a receptor.
Receptor Sequestration: Receptors are internalized and temporarily unavailable.
Receptor Down-Regulation: Receptors are degraded, reducing their numbers on the cell surface.
Receptor Inactivation: Receptors are modified to become less responsive.
Inactivation of Signaling Protein: Some third party protein inactivates a secondary messenger protein
Inhibitory Protein Production: Signaling pathways produce proteins that inhibit receptor activity.
6. What do we mean by tolerance and why can it be dangerous in opioid addiction
Tolerance and Opioid Addiction Progression
Normal State:
Baseline: Receptor activity is regulated by endogenous ligands (e.g., endorphins). Receptor numbers are stable.
Euphoria (Initial drug use):
Increased ligand (opioid) concentration overstimulates opioid receptors, producing euphoria. No tolerance yet.
Tolerance:
With repeated opioid use, receptor downregulation occurs. More ligand (drug) is required to achieve the same effect due to fewer receptors available.
Dependence:
The body adapts to high ligand levels, becoming dependent on external opioids. Endogenous ligands no longer maintain normal function, leading to withdrawal symptoms if opioids are stopped.
Post-Abstinence Risk
Normal After Abstinence:
After stopping the drug, receptor upregulation restores receptor levels to baseline, but endogenous ligand production is still low.
Withdrawal:
Without external ligand (opioid), receptors are understimulated, causing withdrawal symptoms due to insufficient receptor activation.
Recovery:
Over time, the body’s endogenous ligands regain normal function, and receptor activity stabilizes.
Overdose (High Risk):
If opioid use resumes at prior doses, the upregulated receptors are overstimulated by the same ligand concentration, leading to overdose.
Cell Communication - Control of Cell and System Functions
1. Know how Steroids Work
all share common cholesterol backbone
all have a DNA binding domain
even though its a different sequence , this part always does the same thing , just binds to DNA
all have a Hormone binding domain
but these all have a different function
cortisol vs progesterone for instance
Two Families:
Family 1: Includes common steroids like glucocorticoids, androgens, estrogens, progesterones, and mineralocorticoids.
These receptors are cytosolic,
Upon ligand binding, the receptor forms a homodimer and translocates into the nucleus, initiating gene transcription.
Family 2: Includes T3, retinoid acid, RXR, and vitamin D receptors.
These receptors are nuclear
After ligand binding, they form a heterodimer (often with RXR) before binding to DNA and initiating gene transcription.
2. Know the mechanism by which steroid receptors activate new gene transcription
Steroid receptors contain:
Zinc finger motifs in the DNA-binding domain.
These are essential for binding to hormone response elements (HREs) on the DNA.
HRE = nucleotide sequence unique to zinc-finger
regulate the rate of transcription
zink is used as the coordinating ion
The hormone binding to the receptor induces a structural change that releases heat shock proteins, exposes the zinc fingers, and allows the receptor to bind to specific HREs, activating or repressing gene transcription.
Family 1 :
steroid crosses cell membrane
steroid binds to receptor / heat shock protein complex
heat shock proteins dissociate , receptor is activated
receptor moves into nucleus
receptro binds to DNA as a dimer
receptor alters gene transcription , mRNA is made
target protein is made , elicits biological response
Family 2 :
steroid crosses cell membrane
steroid moves into nucleus
steroid binds receptor , activates receptor ( heterodimer )
receptor binds to DNA as heterodimer
receptor alters gene transcription
target protein is made , elicits biological response
3. Know how kinase Receptors Signal
Receptor Tyrosine Kinase Activation :
Binding of ligand causes dimerization
dimerized receptor phosphorylates tyrosine
either auto-phosphorylation or cross-phosphorylation
phosphorylated receptor recruits and activates adapter proteins
kinase cascade
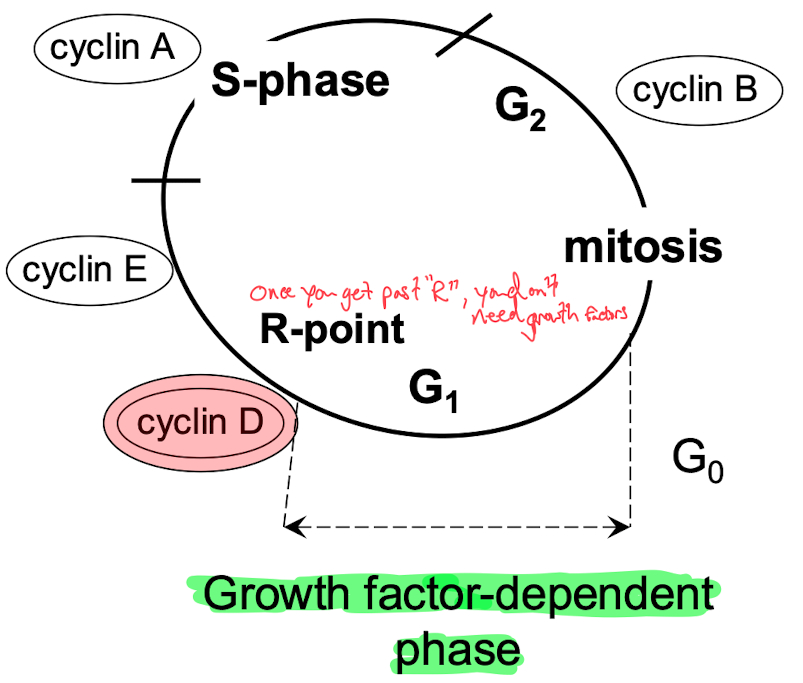
MAP kinases are one of the most common pathways involved in kinase signaling, especially in regulating the cell cycle.
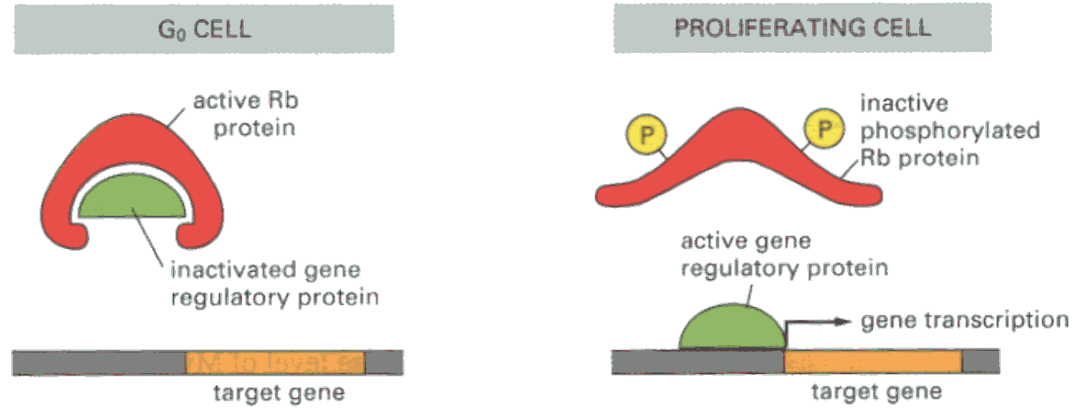
MAP kinases activate CDKs (Cyclin-Dependent Kinases), which in turn regulate cyclins. These cyclins phosphorylate key proteins like RB (Retinoblastoma protein), which regulates whether the cell can progress through the cell cycle. Phosphorylated RB allows progression, driving the cell from G1 into S phase.
Why CDKs and Cyclins Act on the Cell Cycle
They control cell cycle checkpoints, ensuring proper cell cycle progression.
Misregulation (e.g., through oncogene mutations like KRAS or loss of tumor suppressors like RB) can lead to uncontrolled cell proliferation, a hallmark of cancer.
Growth Factor ➡️ MAP Kinase ➡️
Rb = prevents cell cycle progression via inhibition of gene transcription
blocks expression of myc and fos
binds to E2F ( needed to go past R-site )
Phosphorylated Rb allows transition from G1 to S-phase
Misc
"genomic" = traditional = effect is mediated by change in transcription of a gene into mRNA
slow
NISS = nuclear associated steroid signaling
"non-genomic"
fast
effect is mediated by a cell signaling casacade
MISS = membrane induced steroid signaling