BMB 7500 - Exam 2
Chromatin
It is possible to form nucleosomes in the laboratory by mixing purified histones H1 , H2A , H2B , H3 , H4 and 2000 bp linear pieces of purified DNA in high salt (
) and slowly diluting the salt to
Why is it necessary to dilute the salt from
- High salt would block the ionic interaction between histones and DNA phosphates.
Approximately how many nucleosomes will form on each 2000 bp piece of DNA?
- 10 to 13
If histone H1 was omitted from the mixture , would you expect more or fewer nucleosome cores to form on each 2000 bp DNA fragment? Explain why.
Multiple answers were acceptable if justified by clear reasoning
For example :
- more nucleosomes due to more unoccupied linker space
- fewer nucleosomes because H1 stabilizes nucleosome cores
- same number of nucleosomes because H1 does not affect the nucleosome core
If the DNA was in the form of relaxed 2000 bp plasmid circles, would you expect more or fewer nucleosomes to form on each piece of DNA? Explain why.
- Fewer total nucleosomes due to the generation of positive supercoils in the circular DNA
Bacterial Transcription
Explain in molecular terms of the proteins or enzymes that regulate gene expression , what the effect would be ( e.g. increase , decrease , no difference from wild type ) of the following mutations on the expression of the trpA protein :
Deletion of the Trp operator
- increased , due to loss of TrpR binding
Deletion of the Trp leader peptide AUG ( translation start ) codon
- increased , due to loss of 3:4 hairpin attenuation
Deletion of the Trp promoter
- decreased, due to loss of RNA pol binding site
Constitutive ( always ON ) expression of the TrpR gene
- no change, this is the wild type condition
Transfer of the TrpL gene ( 163 bp leader ) to the 3’ end of the Trp operon ( as shown in the figure below )
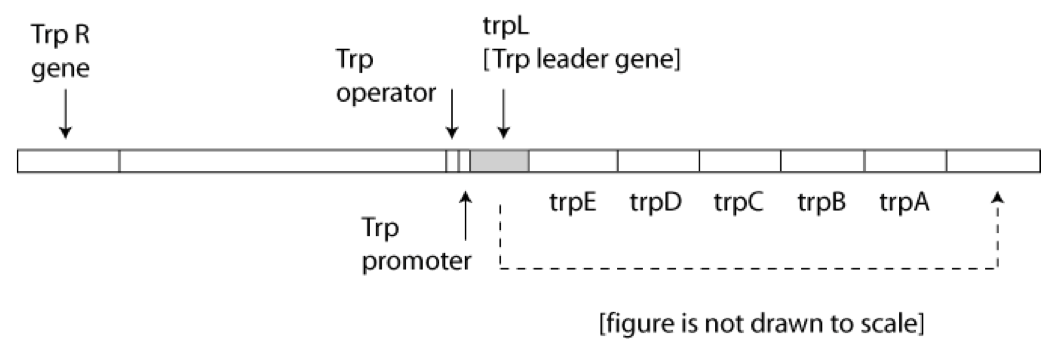
- increased, due to loss of attenuation
Transfer of the TrpR gene from the Trp operon to the position of the LacI gene in the Lac operon
- no effect on Trp operon
Eukaryotic Transcription
Why can’t eukaryotic organisms use attenuation as a mechanism of transcriptional control?
- Ribosomes cannot access the transcribing gene.
Why is it that eukaryotic genes can’t be expressed in bacteria , but bacterial genes can be expressed in eukaryotic?
- Eukaryotic genes contain introns
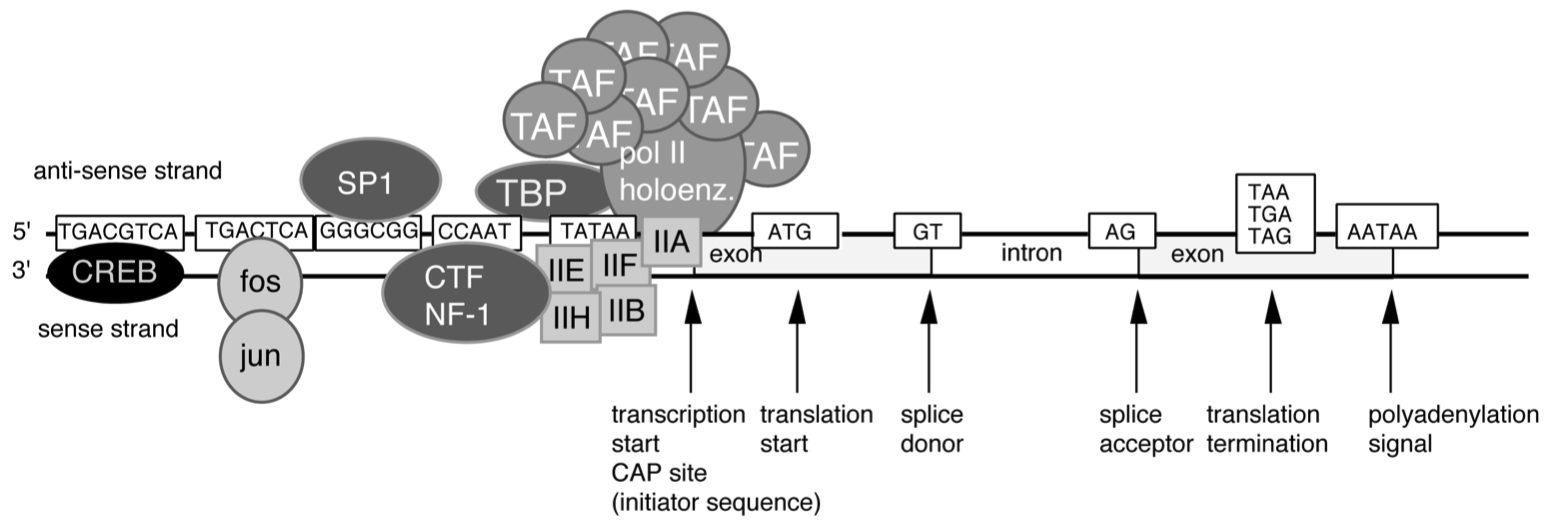
Draw a diagram of a eukaryotic protein coding gene containing three exons and two introns , including the following elements in appropriate spatial relationship to one another :
- 3' untranslated region
- 5' untranslated region
- AATAAA polyadenylation signal
- ‘cap’ site
- binding site for basal transcription factors
- poly-A tail( s )
- splice donor and acceptor site( s )
- translation start site
- TATAAA box
- transcription start site
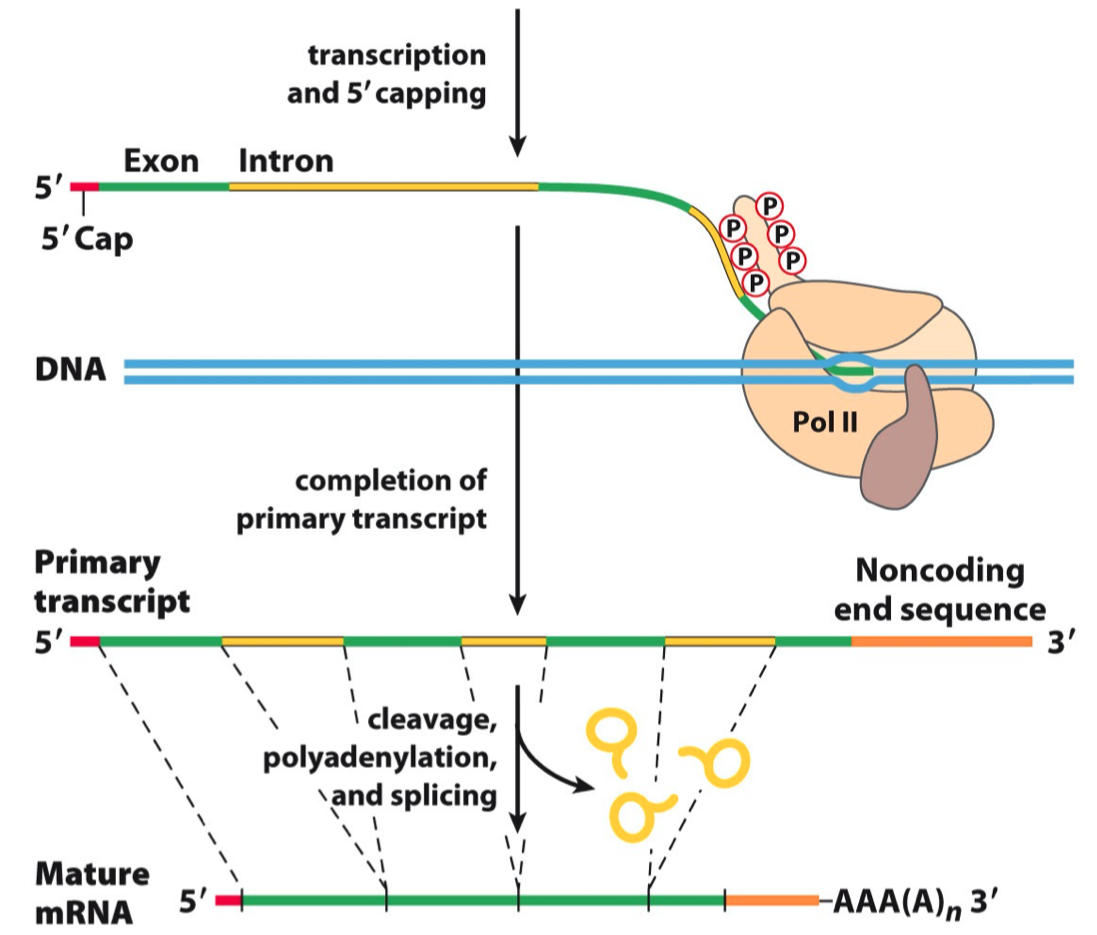
RNA Processing
Each of the major classes of RNAs ( including rRNA , tRNA , pre-mRNA ) undergoes processing before it is transported to the cytoplasm
Mention one processing step in rRNA maturation
- cleavage
Mention two processing steps in tRNA maturation
- Cleavage , CCA addition , base modification
Mention three processing steps in pre-mRNA maturation
- Capping , splicing , poly-adenylation , methylation
DNA Replication
What would be the effect on DNA replication if a bacterial DNA polymerase III mutant had decreased
- Loss of proofreading ability, more errors incorporated into DNA
What would be the effect on DNA replication if a bacterial DNA polymerase I mutant had decreased
- Loss of Okazaki fragment processing and ligation
How is DNA helicase activity different from DNA topoisomerase activity?
- Helicase helps unwind DNA directly ahead of replication fork
- Topoisomerase cuts DNA to remove supercoiling upstream of the replication fork
Which DNA template strand ( leading or lagging ) requires repeated DNA primase activity? Explain why
The lagging strand
because it must be synthesized discontinuously every time the replication fork moves
- because the template strands are antiparallel.
DNA polymerase and DNA ligase can both synthesize phosphodiester bonds. How do their functions differ during bacterial DNA replication?
- DNA polymerase makes a phosphodiester bond when it incorporates a nucleotide
- DNA ligase makes a phosphodiester bond using the energy of ATP or NAD+
What are “Okazaki fragments” and why are they necessary?
- The lagging strand is synthesized discontinuously in small pieces ( Okazaki fragments ) every time the replication fork moves
- They are necessary because the template strands are antiparallel
How do eukaryotic cells avoid re-replication late in
- MCM complexes can only be assembled on origins during G1 phase , and only fired during the following S phase , due to CDK kinase activity.
- They cannot be re-fired again until the cell has gone through mitosis.