BMB 7500 - Exam 1
Nucleotide Synthesis
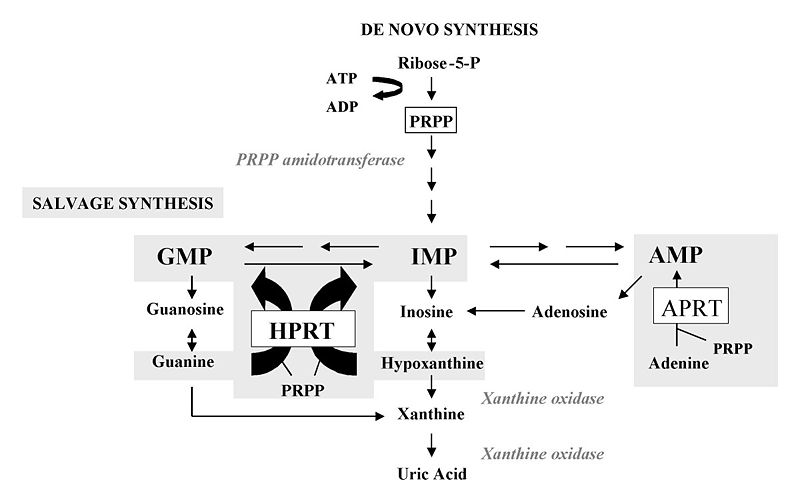
In a broad sense , how does the salvage synthesis of purines or pyrimidines differ from de novo ( "from scratch" ) synthesis ?
- In de novo synthesis , PRPP is formed during the first step
- In salvage synthesis , HGPRTase or APRTase help recycle a ribose-5'-phosphate moiety from PRPP to yield purine nucleotides
Enzymes
What does a DNA ligase enzyme do ?
- Joins two DNA molecules or fragments
What does a reverse transcriptase do ?
- Makes a DNA copy of an RNA molecule
What does HGPRTase do ? ( specify substrates and products )
- Catalyzes the the transfer of ribose-5'-phosphate from PRPP to guanine to form GMP
- Catalyzes the the transfer of ribose-5'-phosphate from PRPP to hypoxanthine to form IMP
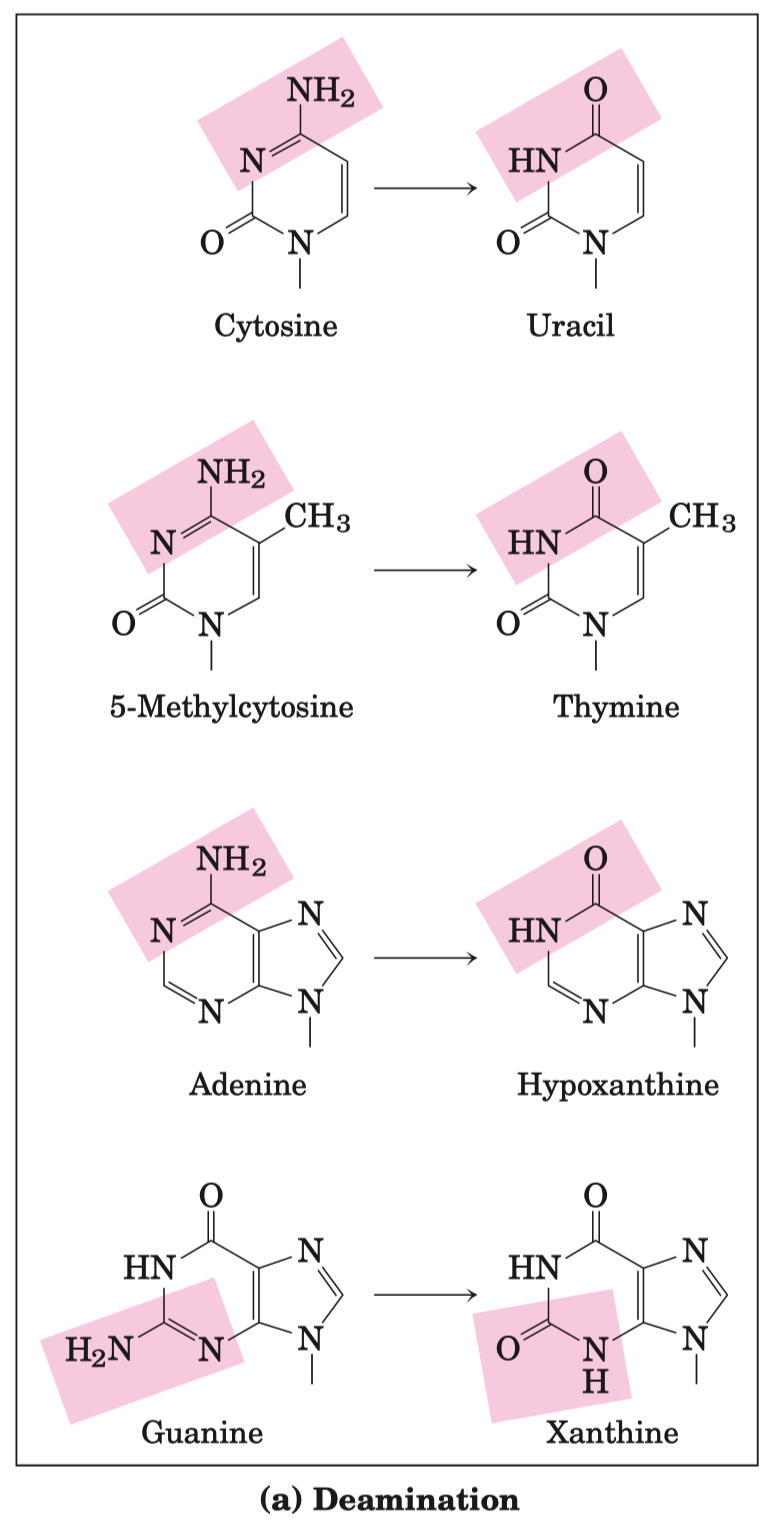
What is the pyrimidine product of ribonucleotide reductase , which does NOT have an exocyclic amino group ? ( be exact about phosphorylation states )
Deoxyribonucleotides, the building blocks of DNA, are derived from the corresponding ribonucleotides by direct reduction at the 2'-carbon atom of the D-ribose to form the 2'-deoxy derivative.
- The reaction is catalyzed by ribonucleotide reductase
- For example , adenosine diphosphate ( ADP ) is reduced to 2'-deoxyadenosine diphosphate ( dADP )
Pyrimidines = Cytosine , Uracil , Thymine
Pyrimidines that don't have an exocyclic amino group = Uracil and Thymine
- Products of ribonucleotide reductase = deoxy-thymidine-tri-phosphate ( dTTP )
Avery , MacLeod & McCarty ( Part 1 )
The paper by Avery et al. argued that DNA is the genetic material.
Describe the observations that led to the conclusions that the genetic information of bacteria is contained in a macromolecule :
- They purified and removed polysaccharide capsule itself , and found the transformation still took place
Describe the observations that led to the conclusions that the genetic information of bacteria is DNA , not RNA , protein , lipid , or carbohydrate :
- Only when they added a DNAase enzyme were they able to STOP the transformation from
- Only when they added a DNAase enzyme were they able to STOP the transformation from
Describe the observations that led to the conclusions that the genetic information of bacteria is inherited :
- All offspring of bacteria that had ben transformed from
- All offspring of bacteria that had ben transformed from
Describe the observations that led to the conclusions that the genetic information of bacteria contains information for the synthesis of macromolecules that are not nucleic acids :
- They
- They
Why did the substance that could transform
- Isotonic ( pure ) water causes the DNA to become unstable
- A low salt solution has free positively charged sodium ions that can help stabilize the negatively charged DNA
Avery , MacLeod & McCarty ( Part 2 )
In the Avery et al. paper , the authors tried to relate the viscosity of the transforming material to its biological activity ( the ability to transform
to bacteria )
Did the authors find high viscosity or low viscosity was related to transforming activity ?
They found DNA was highly viscous
And after exposing DNA to DNAase , it chopped up the DNA and made it less viscous.
- This also destroyed the transforming activity
Explain this relationship specifically in terms of the molecular structure of the transforming material
It suggests the structure needs to remain intact , and that whatever DNAase is doing , makes it such that after its done , DNA can no longer function.
- Said another way , DNA needs to remain in one piece , and does not function if cut up into fragments
Name an enzyme activity , that was NOT used by Avery et al. , that would DECREASE the viscosity of the transforming activity
- Anything that can cut up DNA
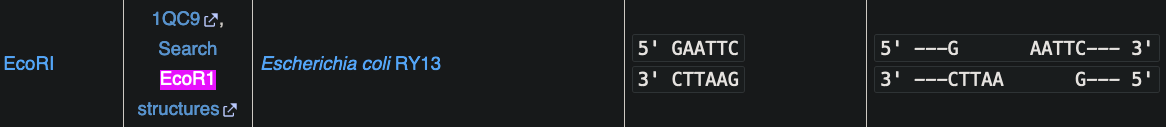
- A restriction enzyme would work such as EcoRI
- The more DNA is cut into fragments , the less viscous it becomes
DNA Structure
What are Chargaff's rules ?
Adenine must pair with Thymine
- Therefore adenine will have the same concentration as thymine
Guanine must pair with Cytosine
- Therefore guanine will have the same concentration as cytosine
To which nucleic acids do Chargaff's rules apply : single stranded DNA , double stranded DNA , single stranded RNA ? Explain
- It only applies to double stranded nucleic acid structures
- Single stranded DNA or RNA isn't forced to pair
What are the two major types of forces that are responsible for B-DNA structure ?
Hydrogen Bonds
Sodium cations stabilize negative charged DNA
the purine base is positioned away from the sugar ( “anti" conformation )
Watson and Crick
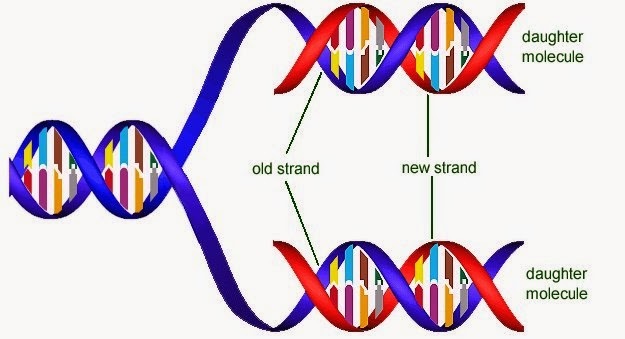
What did Watson and Crick mean by the statement "It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material" ?
- They meant that if the two strands of DNA are separated apart , both of the strands can be used as template for copying the other strand , by complementary base pairing mechanism
Explain in specific molecular detail how the physical structure of the DNA molecule that was proposed by Watson and Crick could explain how the change in
Watson and Crick proposed DNA replicates semi-conservatively
- meaning , each of the two daughter cells will have a copy of their parents DNA
DNA Sequencing
What is this molecule ?
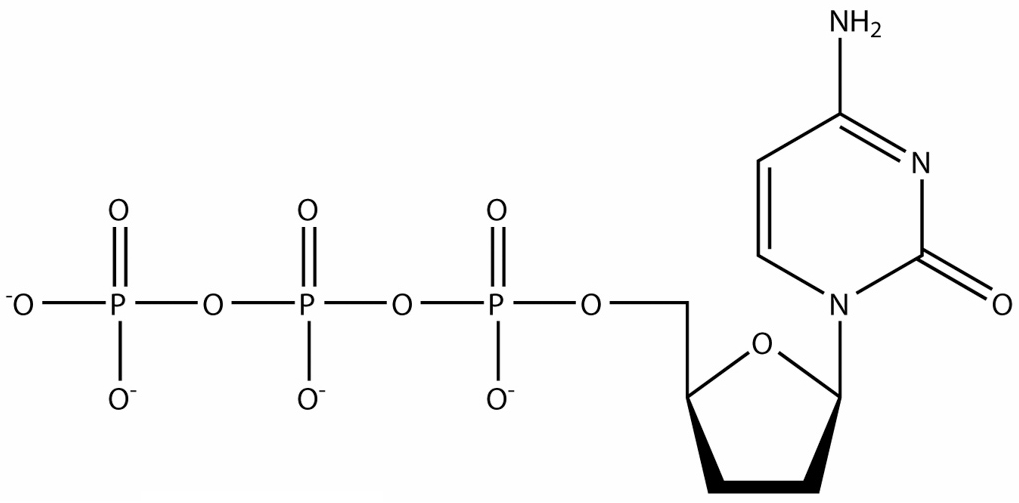
- di-deoxy-cytosine-tri-phosphate ( ddCTP )
Explain in detail how it is used in DNA sequence analysis
- di-deoxy nucleotides are used in Sanger sequencing to cause termination of the growing DNA strand
- every time a cytosine is replicated , the sequence will be terminated and produce variable length fragments
- the sequence can then be calculated by sorting the fragments by their length
DNA Supercoiling
- Ethidium bromiode ( EtBr ) is a flat molecule dye that can insert itself between the base pairs of DNA ( like slipping extra cards into a deck of cards ) without breaking the DNA
- Using gel electrophoresis , a student observes that treating a negatively supercoiled plasmid with increasing concentrations of ethidium bromide results in the plasmid becoming relaxed , and then positively supercoiled.
Using the linking number equation and your knowledge of DNA structure , explain what ethidium bromide must be doing to change the structure of the plasmid DNA so as to result in an increase in its positive supercoiling
- The only way to keep the linking number constant while increasing the
- Its somehow acting similar to topoisomerase and removing negative supercoils

Would the addition of
Z-form DNA has positive supercoils
- Therefore it should INCREASE the likelihood of Z-DNA formation
Restriction Enzymes
Make up a hypothetical double stranded DNA sequence that contains a six base recognition site for restriction enzyme cleavage. ( Label
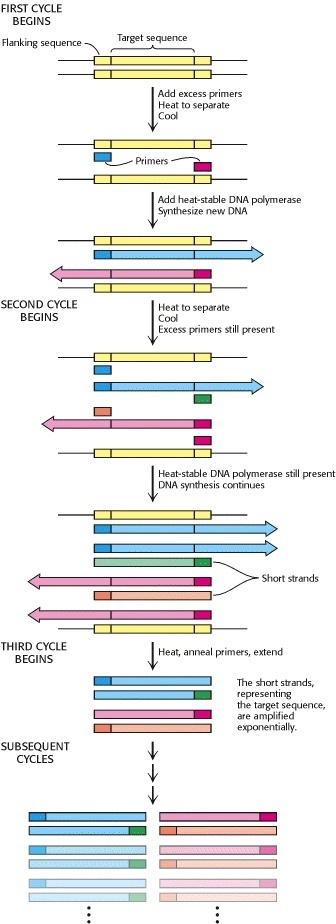
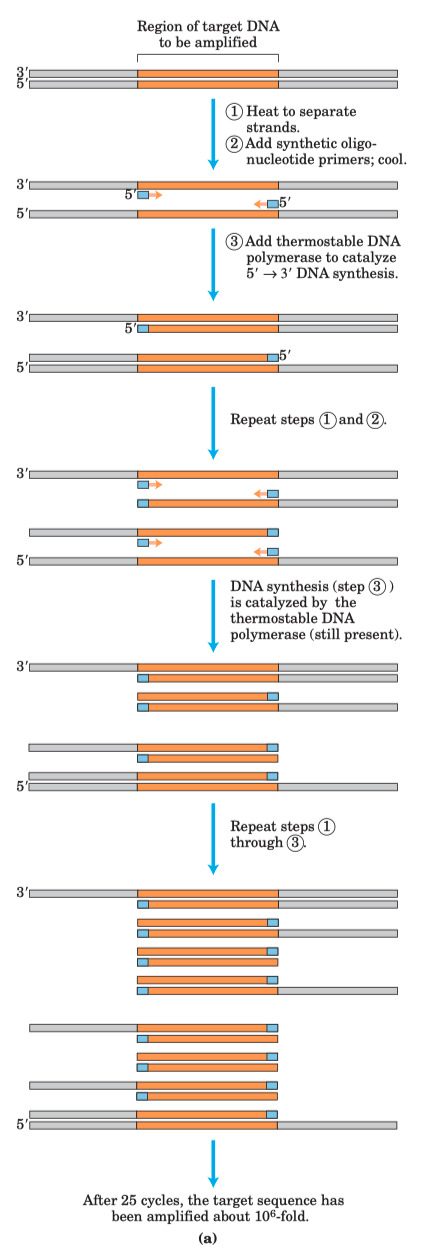
PCR
A student is using the following PCR cycle conditions to amplify a 384 bp segment of DNA :
- Incubate at
for seconds - Incubate at
for seconds - Incubate at
for seconds
Describe what is happening during each step ( 1 , 2 , 3 ) of the PCR cycle
- Step 1 = heated to high temperature to cause denaturation of the double stranded DNA
- Step 2 = primers are added , and DNA is then cooled down to allow the primers to anneal to the separated strands
- Step 3 = solution is then heated up again to allow heat-stable polymerases ( Taq ) to elongate the primers
After gel electrophoresis of the PCR products , the student sees three bands ( 212 bp , 288 bp , 866 bp ) in addition to the desired 384 bp product. Which PCR step ( 1 , 2 , 3 ) should be modified , and how , to try and amplify only the desired 384 bp band ?
- melting temp is directly proportional to percent of perfect matched basepairs
- You can modify the temperature you cool down to in step 2
- If you make it more difficult for the primers to bind by not decreasing the temperature as much in step 2 , then the primers are more likely to bind only to a perfect match
Why must the primers face TOWARDS each other when using PCR to amplify a linear DNA molecule ?
- If they face in opposite directions , you will end up copying the entire length of the plasmid