Exam 4
- Rate is proportional to transition state activation energy = Delta-G-dagger
- Likelihood the reaction occurs = Delta-G of the reaction
- Rate enhancement is proportional to Delta-Delta-G-double-dagger
- Equilibrium is proportional. To Delta-G of the reaction
Catalytic Strategies for Enzymes
- acid / base
Rnase A
Has 2 histidines in the active site
- Histidine at 118 Acts as an acid ( gives proton )
- Histidine at position 12 acts as a base
PH has to be near the
Histidine is a “popular” one for acid / base reactions
Covalent Catalysis
good at binding and stabilizing the transition state
- Lowers activation energy
Metal Ion Catalysis
amino acid chemistry stinks at redox reactions?
- Good at acid base ?
Help with acid base chemistry
- You don’t have to change the pH
Zinc changes the “polarizability” of the water around it
Proximity and Orientation
closer in space = “more probable the bump into each other”
- 5x enhancement
Proper orientation = 100x enhancement
Remove background movement
- Enzyme “calms” the natural vibration of the substrates
- Aka stabilizes the transition state
Geometry
- molecular orbital theory
- Van Der Waal Radii
Problem - Which of the following amino acid residues …
- B , histidine , aspartate , glutamate
Problem 2 - The figure to the left shows a proposed …
Zinc = changes polarizability of water
Arg 145 = helps to bind substrate
During transition state it helps stabilizes through ionic interactions
- Hydrogen bonds
Glu 270 = acts as a base , and then as an acid
Chemical Kinetics
Kinetics helps understand reaction mechanism
Concentration Changes :
- Substrates
- Products
- Intermediates
Concentration Doesn’t Change :
Catalyst
Solvent
Anything with extremely high concentration
- Little change is negligible
Changes in concentration can change the reaction order
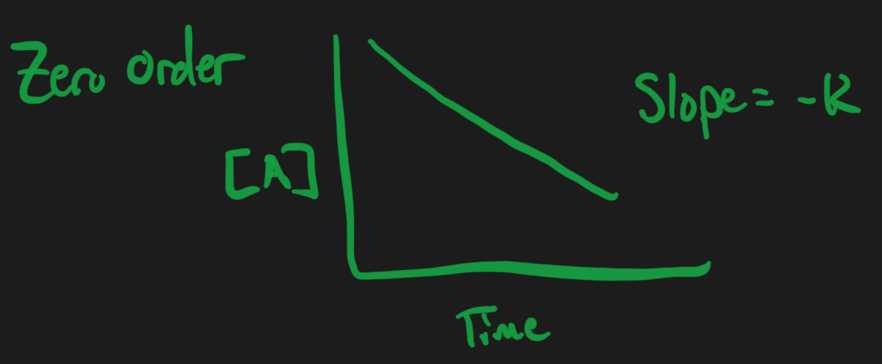
Zero Order Reactions = doesn’t change with respect to concentration
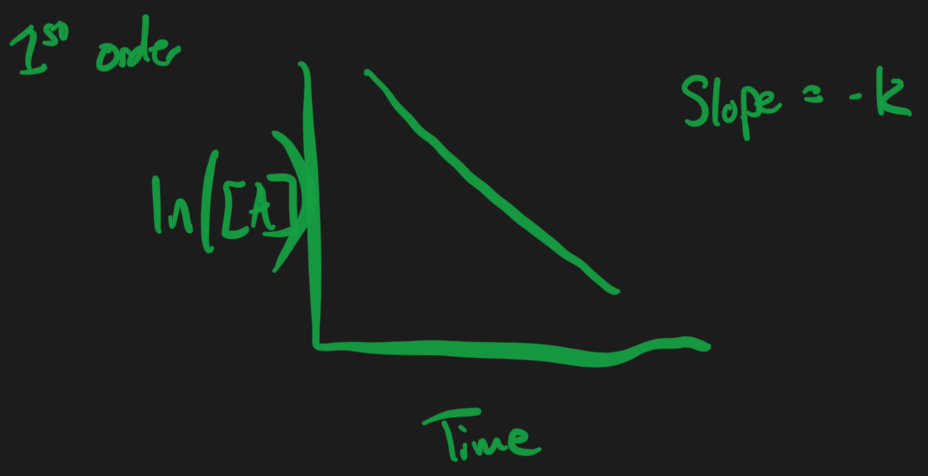
First Order = changes with respect to the concentration of 1 thing
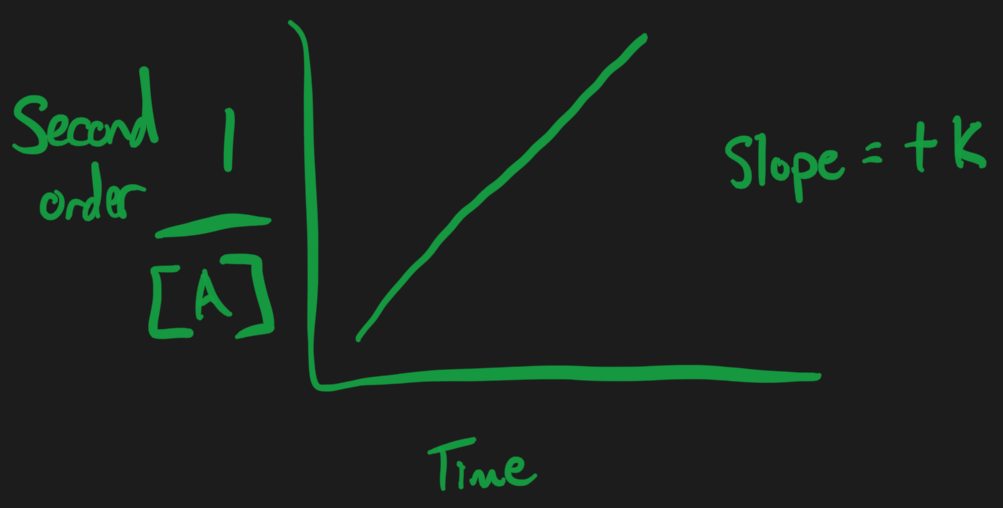
Second Order = changes with respect to the concentration of 2 things
Third Order = ….
Zero Order Reactions
Slope =
Negative because you are subtracting out anything that takes [A] away
- Forward reaction
First Order Reactions
Or
Plot = x-axis = time
- Y-axis =
- Y-axis =
Slope =
Second Order Reactions
Unimolecular :
=
- Or
- Or
Plot = linear
X-axis = time
Y-axis =
Slope =
Bimolecular :
- Harder than unimolecular
- You can view this as change in product formation over time
- Integral Result =
Having 2 things coming together in the proper position and proximity is harder than 1 thing
3 things coming together at the same time is very rare in biology / enzymes
Opposing Reactions
Subtracting anything that takes
- Then multiply by reactions
Add anything that makes it
Multiple by its reactants ??? See transcript @@ 10:48
If you have some 3rd order reaction
But you don’t know which reactant is second order and which reactant is first order
You can add so much concentration of one of the products to create a “pseudo “ order reaction
- Then the rate no longer depends on it
- Makes it “zero” order aka “pseudo” order
Half-Life
- time required to reduce the concentration by half
- Substitute in
- Zero Order =
- First Order =
- Second Unimolecular =
Problems :
Determine the velocity of the reaction :
The reaction has a rate constant of
- Units for
- Half Life =
- Units for
- measures the number of substrate molecules converted to product per enzyme per second when the enzyme is saturated
Transition State and Enzyme Kinetics 1
Transition State Theory = Enzyme binds best to the transition state ??? Ok
K’ = rate of decomposition of transition state to products
Reactants in the transition state are in equilibrium
??? See lecture audio
What is the rate of the reaction proportional to?
- Transition state energy
- Rate
frequency can bump the transition state form products
The equation proves why it is proportional to transition state energy
Enzyme Catalysis
The fastest an enzyme can go is linked to the “fusion”
- How fast can the substrate get to the enzyme
Biggest jump = rate limiting step
How to Lower
- enthalpy has the biggest effect
Imaginary Stickase
“picture” of transition state theory
If it prefers to bind to the transition state , then it lowers the energy of activation
We need to have the number of molecules to reach the transition state to be high
- We can use temperature to help
- have to overcome peak of transition state
- Higher temperature has more molecules able to reach transition state
Enzymatically , we only have 1 line , because the body does not alter temperatures
Enzymes cause left shift on the curve
- Kinetic energy vs number of molecules graph on the right
Mechaelis-Menten
not the best , because of assumptions
Assumes there is only 1 substrate
Zero-order kinetics , doesn’t rely on the concentration of anything
- This doesn’t accurately depict the situation in biology
- But gives a good enough approximation to compare and conceptualize
- Bunch of rate constants on each equilibrium position
Velocity of product formation =
[ES] is a function of other things
There are a couple assumptions you can make
Assume an equilibrium expression
Textbook says it doesn’t work , but you can make this assumption but math is more complicated
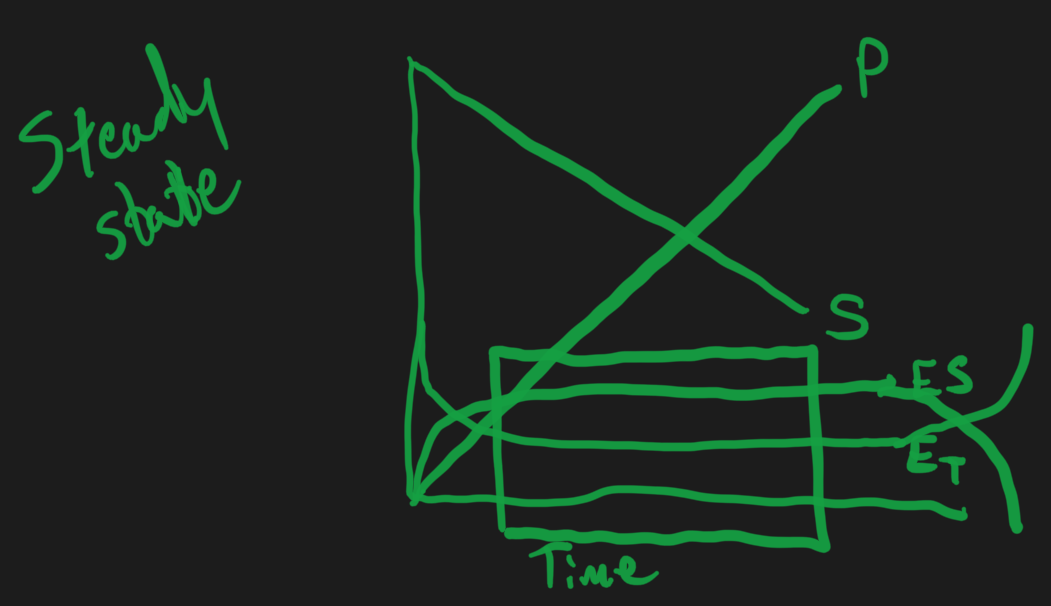
But instead we use the steady state approximation
The change in enzyme-substrate concentration with respect to time is zero
Only works if we are operating under “saturation” conditions
Start with really high concentration of substrate , binds to the enzyme
As soon as the enzyme releases substrate , new substrate binds immediately
- there is so much substrate around , there there is no chance of the enzyme ever being by itself
Total amount of enzyme = free enzyme concentration + enzyme-substrate concentration
Set
Then some other step :
High
Low
- Equals substrate concentration at
- Equals substrate concentration at
Now “substitute that in , and solve for [ES]” , ok
Now plug this into the original :
- Getting rid of K and K-minus-one
If operating under zero-order kinetics : The only thing that dictates
Why does
- Asdfasf see transcription
- Enzyme’s affinity for substrate dictates rate ,
Catalytic Efficiency =
- “Rate / affinity “
Line-Weaver Burk Plot
Double-Reciprocal Graph
Graph of kinetic data
- Substrate concentration vs velocity
Line-weaver is a “normalization” , creates a linear curve
By plotting it this way , we can easily determine variables
Slope =
Y-Intercept =
- Upward Shift on the Y-Axis = smaller
- Downward Shift on the Y-Axis = larger
- Upward Shift on the Y-Axis = smaller
X-Intercept = Set Y = 0 , solve for X
- Left Shift on the X-Axis = smaller
- Right Shift on the X-Axis = larger
Better because of Pipetting reasons ?
Problem : An enzyme catalyzed reaction has a
- as long as substrate and enzyme are in the same units , you don’t have to convert
Kinetics and Mechanisms
3 major reaction mechanisms
random substrate binding
- intersect at y-axis
ordered /sequential substrate binding
- first enzyme causes conformational change that is needed for the second substrate to bind
ping-pong mechanism
bind to substrate and release product
during this step , you modify the enzyme
- the modified enzyme-substrate complex can then bind to second substrate
Lineweaver-Burk Plot
Slope =
Y-Intercept =
- Upward Shift on the Y-Axis = smaller
- Downward Shift on the Y-Axis = larger
- Upward Shift on the Y-Axis = smaller
X-Intercept = Set Y = 0 , solve for X
- Left Shift on the X-Axis = smaller
- Right Shift on the X-Axis = larger
Inhibitors
Competitive Inhibitors
all 3 are reversible
binds to the same place as the substrate
increases
$V_{max} stays the same
cross at y-axis
these equations help us in drug design
- trying to inhibit a target , use these equations to prove / demonstrate viability
alcohol dehydrogenase has multiple different substrates it can use
ethanol , methanol , propanol
- whichever is lower = higher affinity = the winner of which binds
Uncompetitive Inhibitor
Binds to ES complex
means
binds in a different position
has to be enough substrate for enzyme to bind to form ES complex , if there is no ES complex , the inhibitor can’t bind
- Not enough substrate for it to affect
competitive inhibitor is effective a low substrate concentrations
uncompetitive is effective at high substrate concentrations
perfect drug design is uncompetitive , because it can never be beaten
most of the time though drugs are competitive
rare in mechaelis-menten kinetics
- less rare when there is 2 substrates
Prime = denotes inhibitor is binding to ES
lines are parallel
Mixed Inhibitor
binds to the enzyme or enzyme-substrate complex
has a measurable
subset of mixed called “non-competitive”
Non-Covalent Inhibitors :
Competitive = Binds Enzyme
- effective at low substrate concentrations
- LWB = crosses @
Uncompetitive = Binds ES complex
- effective at high substrate concentrations
- LWB = does not cross
Mixed = binds E or ES
- effective at low and high concentrations
- one subset called “non-competitive” , where
- LWB = crosses anwhere but at the
SEE TRANSCRIPT FOR WHAT NOT TO MISS !!!
- mixed
- mixed
- mixed
- non-competitive
Problem : An enzyme has a
Problems Slide Deck
#2 on homework 6 = Methanol Question with the dog
- substrate =
- inhibitor =
- we wan’t to make sure its reduced to
- make substitutions and then algerbra
A first order reaction is
solve for
- substitute this in
- something cancels
For a simple reaction the
- Rate Enhancement =
- Rate Enhancement =
From the following date determine
scatter plot ( 1 / S ) and ( 1 / V ) in excel
find equation of a line
Now we can find Vmax and Km
- same units on
- same units on
set equation equal to 0
- same units as substrate concentration
From the following data determine :
The type of inhibitor
plot everything as inverses
- ( 1 / S ) and ( 1 / V )
- should be positive slope , straight line
bottom line =
the middle line =
top line =
lines are not parallel , slopes are different
- rules out uncompetitive
where do they cross ?
- the intercepts are different by only 0.01 = competitive
Apparent
No Inhibitor :
y = 0.4843x + 0.1951 ; find
y = 0.755x + 0.1969 ; find
y = 1.0062x + 0.1861 ; find
uncompetitive uses alpha-prime
alpha or just
Estimate
- experimental error
- lineweaver-burk plot is an estimation , creates error
The catalytic efficiency of many enzymes depends on pH. Chymotrypsin shows a maximum value of
amino acids responsible for
- 8 and 10 = probably lysine
An enzyme follows simple Michaelis-Menten kinetics
- enzyme preferentially binds to
- Cat Eff =
- Cat Eff =
Draw curves that would be obtained when velocity vs
Initial rate data for an enzyme that obeys Michaelis-Menten kinetics……
- y-intercept =
Misc
Office Hour - Next Week :
- Monday = 10:00 - 13:00
- Tuesday = 09:00 - 12:00
Final Exam :
- 35% from Dr. Leffak
- Exam 3 = high level overview
- Exam 4 = practice exam will be posted
- Time = 10:15 - 12:15
Enzyme Examples
Chymotrypsin and lysozyme ?
Chymotrypsin :
protease , cleaves peptide bonds. Used in sequencing
cleaves after an aromatic on the C-terminus side
poster child for how cool enzymes are
Catalytic Triad = Aspartate 102 , Histidine 57 , and Serine 195
Specificity Pocket = where the amino acid that
scissored bond = bond that is cleaved
chymotrypsin vs trypsin
trypsin doesn’t have the serine in the core of the pocket
- its swapped out for aspartate 189
the serine doesn’t disrupt bonding in the aromatic group
glycine is important because it is flexible and it is small enough to allow the aromatic group to fit into the specificity pocket
Trypsin cleaves after a positive amino acid
- so it has a negative charged amino acid in the core of the pocket
elastase cleaves after small , neutral amino acids
- has a neutral core , with bulky amino acid side chains. Only allows small amino acids like alanine to fit inside
Serine Proteases ( serine in active site ) = Chymotrypsin , trypsin , elastase
have 40% of their sequences is identical
specificity pocket is unique
clotting cascade = all serine proteases ( really common mechanism )
produced by pancreas , secreted into G.I track
produced and stored as the inactive ( pro ) form
then something is cleaved off to produce the active enzyme
= enzyme control : proteolytic cleavage
- aka a zymogen
Figure 6-22 : Flow Charge
what are the 3 amino acids in the catalytic triad doing ?
- Aspartate 102 = has polarizing effects on the histidine
- Histidine 57 = acting as a base
- Serine 195 =
Mechanism :
serine attacks sicissle bond
this transfers a proton to the histidine
- the ability to transfer this proton is aided by the polarizing effects of Aspartate 102
this forms a tetrahedral intermediate
the intermediate decomposes
proton donation
this forms the acyl enzyme intermediate
- this pushes part of the intermediate into the oxyanion hole
leaving group leaves
water is added
reverse of 1st step
- has to return to original form to be an “enzyme”
Reasons Serine Proteases are Classical Examples
- Because of the attack and forming a tetrahedral intermediate , it forms a covalent bond
- “transfer of proton” = acid base chemistry
- oxyanion hole = “preferentially binding to transition state”
during formation of intermediate , we are changing how it sits in the enzyme
pushed into the oxyanion hole
- then it hydrogen bonds here
the best enzymes preferentially bind to the transition state
Figure 11-29
Hydrogen bonding an polarizing effects matter
hydrogen bonding to the “Asp”
some hydrogen bonds are stronger than others
low barrier hydrogen bond :
the nitrogen on the histidine and the carboxylic acid on the asp ,
the hydrogen is shared between the two amino acids
- less as a “hydrogen bond” , more as a “shared hydrogen”
- makes the bond stronger ( think covalent bond electron sharing )
If Asp is mutated and swapped for Glycine , what happens to
it decreases , catalytic rate goes down
If it is instead replaced with Glu , it would be more unpredictable
you still have carboxylic acid to form hydrogen bond
but now its 3d space problem
- there is a chance it lines up better and increases
- there is a chance it lines up better and increases
which affects
serine or aspartate ?
- the serine , because the serine is the main player in the mechanism
Lysozyme
found in mucous membranes , lacrimal glands
attacks carbohydrate portion of cell membranes in bacteria
specifically binds to 6 residues
- cleaves between
- cleaves between
similar to chymotrypsin
C-2 Fluorine = bad leaving group , won’t allow reaction to occur
substrate is “locked in place” = allows researcher to determine structure of the enzyme
- without the substrate their, its harder to pinpoint which are the “catalytic residues”
Catalytic Mechanism :
2 paths because historically their are two different research groups with different conclusions for how the mechanism actually works
Left Mechanism :
Glu = acts as an acid ( donates proton ) , then in the next step it acts as a base ( takes hydrogen from water )
- then we have a break in carbohydrates
Asp = sitting there chilling , polarizing the electrostatic interaction
Right Mechanism :
- Glu = acting as an acid in first step. In the next step , the asp forms a covalent bond in the transition state
Right side = the winner ?
- proved via fluorine ?
Lysozyme Activity vs pH
catalytic mechanism is dependent on two ionizable side chains
- asp needs to be deprotonated
their pKas are pretty similar
using titration of the enzyme , the pKa of the Glu = 6.2 = higher than it is in the table in the book
- surrounding environment alters pKa values
pKa of Asp 3.7
allows for wider range
when it is acidic < 3.7 = non active = protonated = can’t act as a nucleotide
at high pH > 6.2 , the Glu will donated a proton = can’t act as an acid anymore
delicate balance [ 3.7 , 6.2 ] = most active environment for enzyme = highest
Problem - Predict the effect of mutating ASP 102 of trypsin on substrate binding and catalytic activity
- if mutate Asp 102 , catalytic activity decreases . Binding is harder to predict.
Lysozyme residues Asp 101 ….
- they are part of a hydrogen bonding network
- with Asp is sitting right there
- others are part of a larger hydrogen bonding network
- if you mess up the network , you mess up the chemistry
Are we going to effect enzymes near equilibrium or far from equilibrium ?
- trying to effect this pathway
- both enzymes are far from equilibrium
- if they are both far from equilibrium , and you are trying got control , then turn one of the valves……
Final Exam Study Sessions
pH range around 6.0
steady state approximation
Uncompetitive inhibition , line-weaver burk plot = parallel lines
with inbititor = x-axis-left-shifted =
Competitive Inhibition :
With Inhibitor = x-axis-right-shifted :
Practice Test - #7 :
enzyme is active in 6.5 to 8.5
competitive inhibitor can only bind at 6.5
at pH of 8.5 , the competitive inhibitor is deprotonating , and then no longer able to interact with the enzyme
- unlikely enzyme would be deprotonating
Competitive Inhibitor :
- once you find K_m^{app} , then find alpha
- solve for alpha
- then you use th equation :
- then solve for
Practice Test - #3 :
- Rate Enhancement =
- Rate Enhancement =
- Rate Enhancement =
Practice Test - #4 :
normally it has a negative charge
if we replace it with a positive charge , the
Homework 6 - Question #2 :
Velocity of Uninhibited :
Get rid of 95%
Substrate Concentration
Ethanol = Inhibitor , 10 mM = 0.01 M
Inhibitor Concentration =